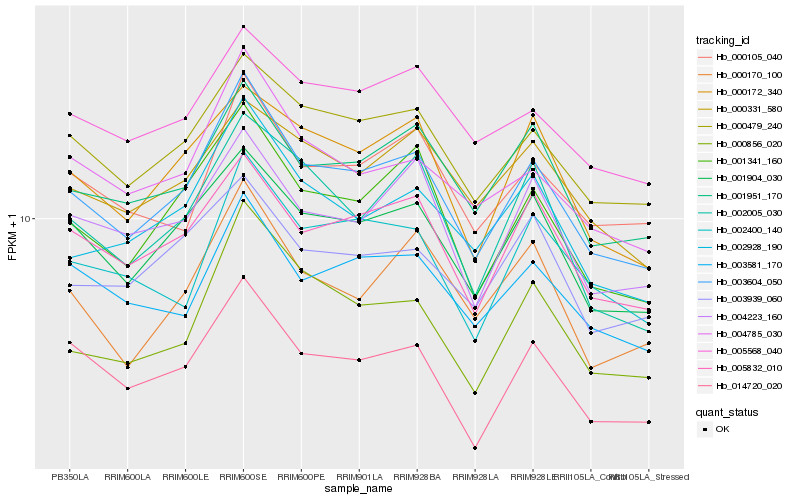
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003604_050 |
0.0 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001904_030 |
0.0555536546 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 3 |
Hb_001341_160 |
0.0565305616 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000479_240 |
0.0584158492 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005832_010 |
0.0634570001 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002928_190 |
0.0655752401 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 7 |
Hb_004785_030 |
0.0677034056 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 8 |
Hb_001951_170 |
0.0678003566 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 9 |
Hb_014720_020 |
0.0696853166 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 10 |
Hb_003581_170 |
0.0716525054 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004223_160 |
0.0737519315 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000172_340 |
0.0738688938 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 13 |
Hb_000856_020 |
0.0741014362 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 14 |
Hb_000105_040 |
0.0744769442 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 15 |
Hb_000170_100 |
0.0772121597 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 16 |
Hb_005568_040 |
0.0772857997 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002400_140 |
0.0784087745 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 18 |
Hb_003939_060 |
0.0791249998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002005_030 |
0.0795178009 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 20 |
Hb_000331_580 |
0.0808746674 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |