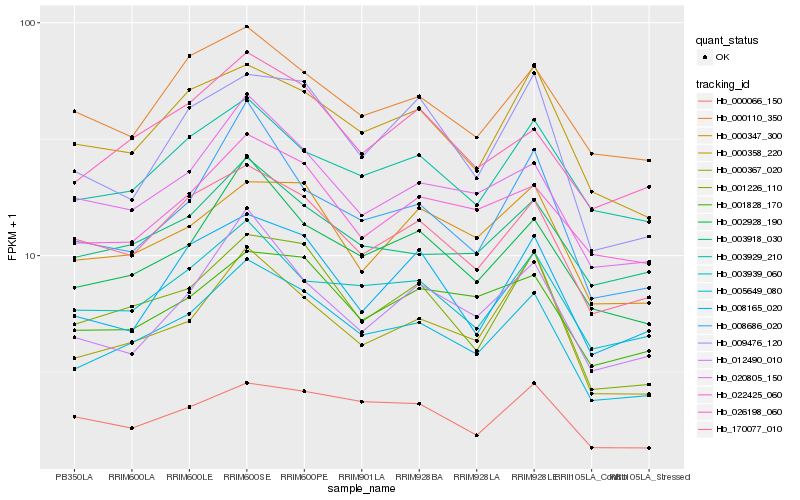
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005649_080 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 2 |
Hb_002928_190 |
0.0623460755 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 3 |
Hb_001226_110 |
0.0638216325 |
- |
- |
PREDICTED: uncharacterized protein LOC105638106 [Jatropha curcas] |
| 4 |
Hb_170077_010 |
0.0695129266 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 5 |
Hb_020805_150 |
0.0725048387 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_000358_220 |
0.0736100244 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 7 |
Hb_003939_060 |
0.0758819043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_026198_060 |
0.0760005794 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 9 |
Hb_000367_020 |
0.0778337487 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 10 |
Hb_003929_210 |
0.080339699 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 11 |
Hb_022425_060 |
0.0804155849 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 12 |
Hb_003918_030 |
0.0807869034 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001828_170 |
0.0815874585 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 14 |
Hb_000347_300 |
0.0835169946 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 15 |
Hb_000110_350 |
0.0860111385 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 16 |
Hb_008686_020 |
0.086877567 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 17 |
Hb_012490_010 |
0.0873408897 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 18 |
Hb_008165_020 |
0.0877422872 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000066_150 |
0.0891360242 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 20 |
Hb_009476_120 |
0.0892465963 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |