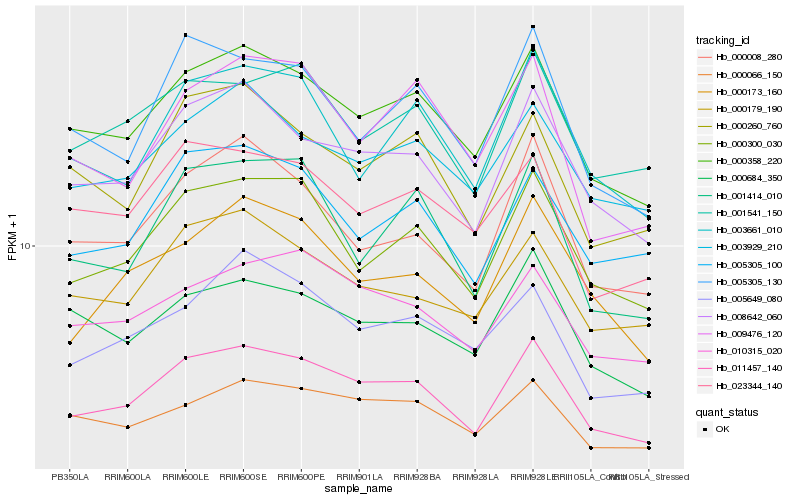
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011457_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649947 [Jatropha curcas] |
| 2 |
Hb_000358_220 |
0.054452005 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 3 |
Hb_008642_060 |
0.0570572685 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 4 |
Hb_000300_030 |
0.0668355265 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 5 |
Hb_000008_280 |
0.0823509117 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 6 |
Hb_010315_020 |
0.082398905 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 7 |
Hb_000173_160 |
0.0825206392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000066_150 |
0.0828301912 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 9 |
Hb_001541_150 |
0.0835882664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001414_010 |
0.0838419443 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 11 |
Hb_005305_130 |
0.0838445027 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 12 |
Hb_003661_010 |
0.0867203851 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005305_100 |
0.0870301551 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003929_210 |
0.0880226144 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 15 |
Hb_000260_760 |
0.088395511 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 16 |
Hb_005649_080 |
0.0892725647 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 17 |
Hb_000684_350 |
0.0901198595 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009476_120 |
0.090710067 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 19 |
Hb_000179_190 |
0.0909605513 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 20 |
Hb_023344_140 |
0.0911962887 |
- |
- |
PREDICTED: WD repeat-containing protein 70 [Jatropha curcas] |