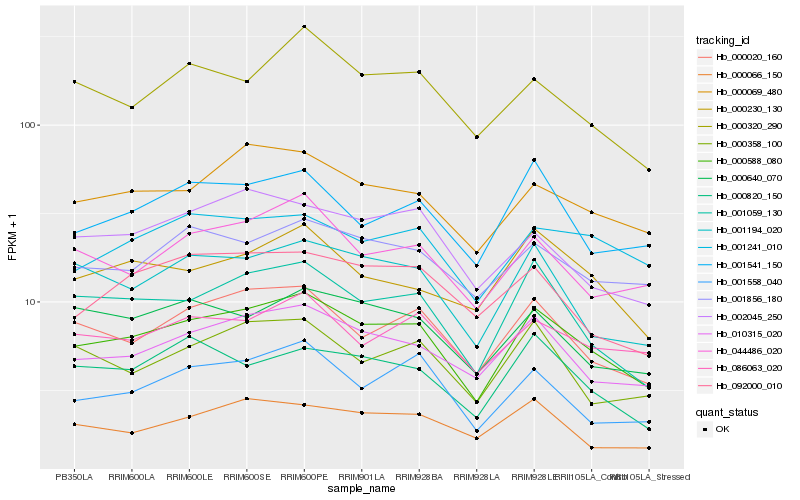
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000588_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 2 |
Hb_010315_020 |
0.0715274172 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 3 |
Hb_001059_130 |
0.0770391597 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000020_160 |
0.0772547955 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 5 |
Hb_000069_480 |
0.0811167062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001194_020 |
0.0843407016 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 7 |
Hb_044486_020 |
0.088905045 |
- |
- |
CASTOR protein [Glycine max] |
| 8 |
Hb_001856_180 |
0.0889489805 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 9 |
Hb_001558_040 |
0.0906915956 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 2 [Jatropha curcas] |
| 10 |
Hb_001541_150 |
0.0908105853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000320_290 |
0.0940817958 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 12 |
Hb_086063_020 |
0.0947174537 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 13 |
Hb_000230_130 |
0.0954805263 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 7 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000820_150 |
0.095707941 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 15 |
Hb_001241_010 |
0.0957922587 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000640_070 |
0.0959920736 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 17 |
Hb_000358_100 |
0.0961130077 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 18 |
Hb_000066_150 |
0.0979493656 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 19 |
Hb_002045_250 |
0.0980081463 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 20 |
Hb_092000_010 |
0.0981457893 |
- |
- |
kelch repeat-containing F-box family protein [Populus trichocarpa] |