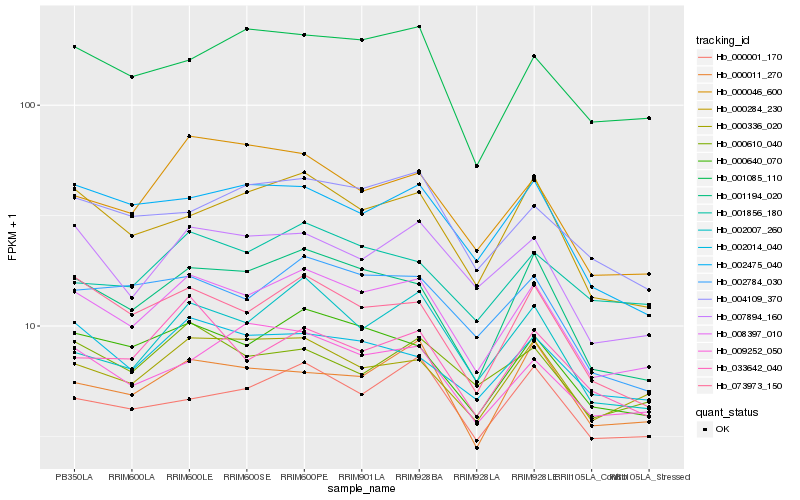
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008397_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000640_070 |
0.0580129854 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 3 |
Hb_001194_020 |
0.0594851768 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 4 |
Hb_000336_020 |
0.0660649789 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 5 |
Hb_000284_230 |
0.0676271638 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 6 |
Hb_073973_150 |
0.0676281018 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007894_160 |
0.0693215907 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 8 |
Hb_002784_030 |
0.0718056972 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 9 |
Hb_002014_040 |
0.0745718937 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 10 |
Hb_001085_110 |
0.0755816073 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 11 |
Hb_002007_260 |
0.075747827 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 12 |
Hb_000001_170 |
0.0774490186 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_000011_270 |
0.0774713436 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 14 |
Hb_004109_370 |
0.0775339264 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 15 |
Hb_002475_040 |
0.0778544869 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 16 |
Hb_009252_050 |
0.0790274903 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_033642_040 |
0.0803699458 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000610_040 |
0.0807285284 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001856_180 |
0.0809619407 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 20 |
Hb_000046_600 |
0.0812588926 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |