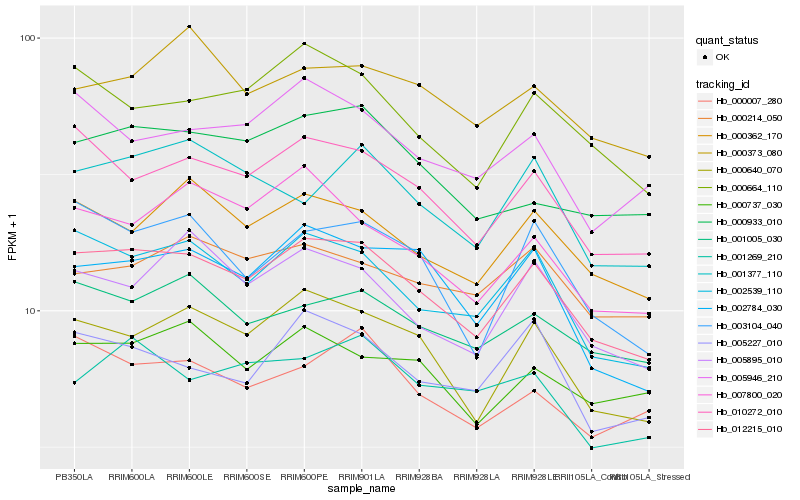
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012215_010 |
0.0 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 2 |
Hb_002539_110 |
0.0533426114 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 3 |
Hb_010272_010 |
0.054223013 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 4 |
Hb_000640_070 |
0.0546379712 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 5 |
Hb_000362_170 |
0.0630213235 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 6 |
Hb_000664_110 |
0.0675314158 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 7 |
Hb_001005_030 |
0.0701415878 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 8 |
Hb_002784_030 |
0.0703107235 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 9 |
Hb_005895_010 |
0.0705823994 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003104_040 |
0.07195916 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 11 |
Hb_000737_030 |
0.0747770106 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005227_010 |
0.0756233192 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000933_010 |
0.0759630338 |
- |
- |
- |
| 14 |
Hb_005946_210 |
0.0762183643 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 15 |
Hb_007800_020 |
0.0775229598 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000007_280 |
0.0776318805 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 17 |
Hb_001377_110 |
0.0798935259 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 18 |
Hb_000373_080 |
0.0803090854 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 19 |
Hb_000214_050 |
0.0812278957 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001269_210 |
0.0818602559 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g76280 isoform X1 [Jatropha curcas] |