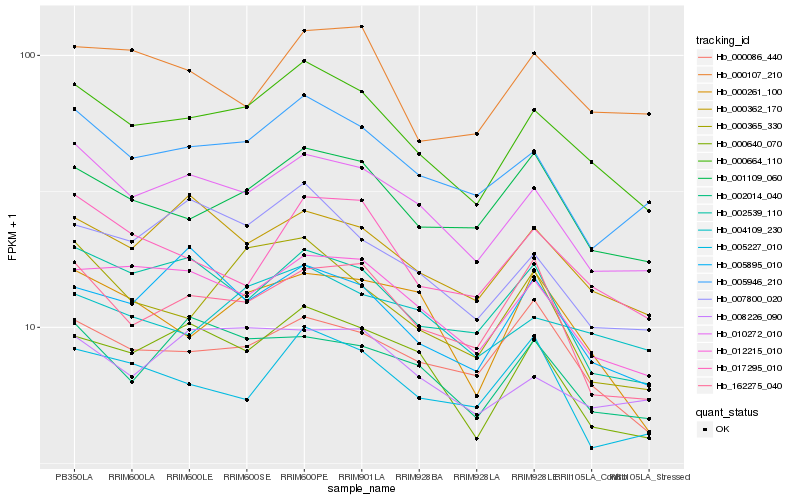
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000664_110 |
0.0 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 2 |
Hb_010272_010 |
0.059523049 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 3 |
Hb_012215_010 |
0.0675314158 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 4 |
Hb_005946_210 |
0.0691607285 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 5 |
Hb_000640_070 |
0.0718383029 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 6 |
Hb_001109_060 |
0.0729009655 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_000362_170 |
0.0769929489 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 8 |
Hb_017295_010 |
0.0782694507 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 9 |
Hb_007800_020 |
0.079403565 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004109_230 |
0.0794139652 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_008226_090 |
0.0822153026 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000365_330 |
0.0823152984 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_002539_110 |
0.0826162569 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 14 |
Hb_000086_440 |
0.0831658351 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 15 |
Hb_002014_040 |
0.0860530488 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 16 |
Hb_162275_040 |
0.0874731415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005895_010 |
0.0876106034 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000261_100 |
0.0894760807 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000107_210 |
0.0897579482 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 20 |
Hb_005227_010 |
0.0901953442 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |