| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
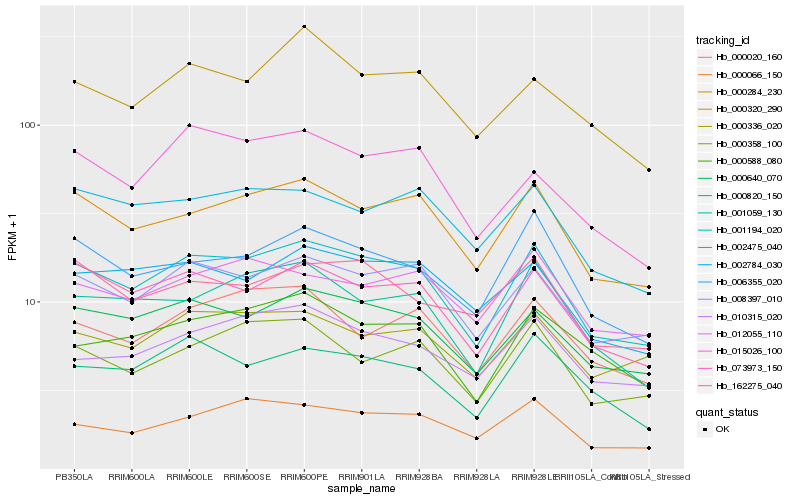
| 1 |
Hb_001194_020 |
0.0 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 2 |
Hb_008397_010 |
0.0594851768 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000284_230 |
0.0596484158 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 4 |
Hb_000640_070 |
0.0612187503 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 5 |
Hb_073973_150 |
0.0629693934 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006355_020 |
0.076491514 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 7 |
Hb_001059_130 |
0.0776469104 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000820_150 |
0.0787428053 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 9 |
Hb_000358_100 |
0.079200104 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 10 |
Hb_000066_150 |
0.0821100168 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 11 |
Hb_002784_030 |
0.0839332226 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 12 |
Hb_000588_080 |
0.0843407016 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 13 |
Hb_010315_020 |
0.0852734961 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 14 |
Hb_002475_040 |
0.0863180323 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000020_160 |
0.0865816019 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 16 |
Hb_000336_020 |
0.0870932775 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 17 |
Hb_012055_110 |
0.0901661612 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 18 |
Hb_162275_040 |
0.0910821195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_015026_100 |
0.0915612542 |
- |
- |
PREDICTED: transmembrane protein 184C-like [Jatropha curcas] |
| 20 |
Hb_000320_290 |
0.0922965656 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |