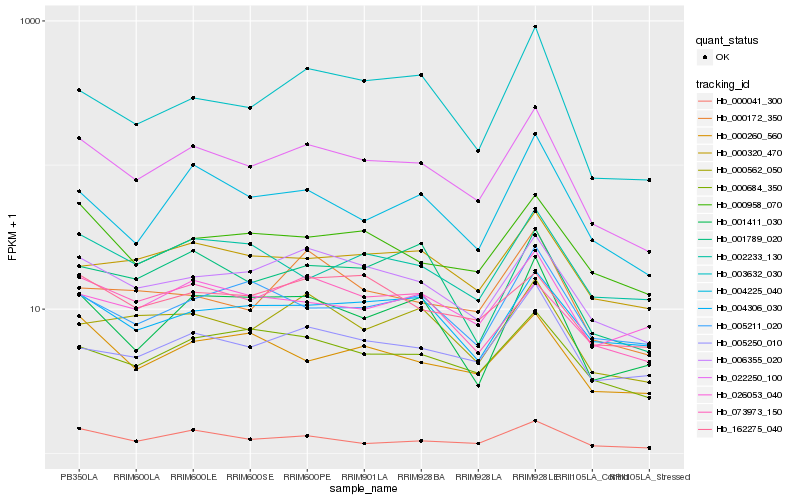
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022250_100 |
0.0 |
- |
- |
calnexin, putative [Ricinus communis] |
| 2 |
Hb_006355_020 |
0.0726833504 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 3 |
Hb_000041_300 |
0.0869188407 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized ATP-dependent helicase C23E6.02 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000684_350 |
0.0960476561 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000958_070 |
0.0991003443 |
- |
- |
hypothetical protein PRUPE_ppa007681mg [Prunus persica] |
| 6 |
Hb_005250_010 |
0.0999086662 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 7 |
Hb_000562_050 |
0.1010395205 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 8 |
Hb_003632_030 |
0.1014285683 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 9 |
Hb_004225_040 |
0.1022390949 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Populus euphratica] |
| 10 |
Hb_000320_470 |
0.1047091644 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 11 |
Hb_004306_030 |
0.1050741321 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 12 |
Hb_001411_030 |
0.1061240907 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 13 |
Hb_026053_040 |
0.106889781 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001789_020 |
0.1075842206 |
- |
- |
PREDICTED: tripeptidyl-peptidase 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_162275_040 |
0.1111986781 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002233_130 |
0.1128043451 |
- |
- |
transporter, putative [Ricinus communis] |
| 17 |
Hb_000260_560 |
0.1130089786 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 18 |
Hb_073973_150 |
0.1131315501 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000172_350 |
0.1133062122 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 20 |
Hb_005211_020 |
0.113453382 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |