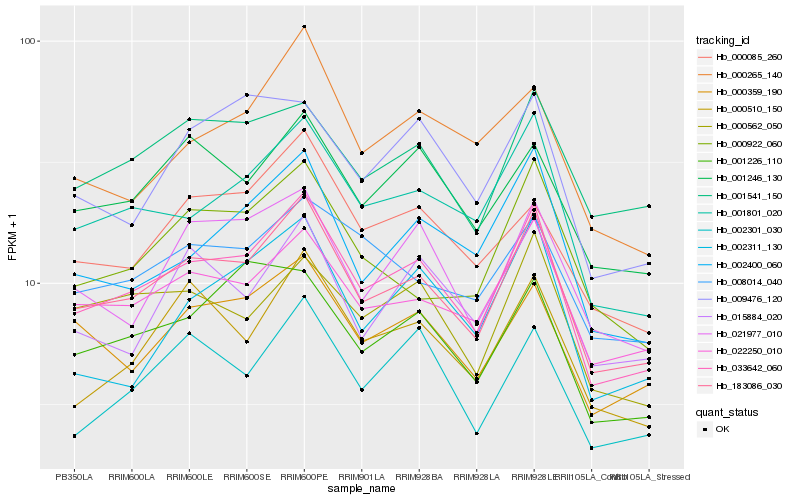
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033642_060 |
0.0 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_183086_030 |
0.0311578932 |
- |
- |
GTPase-activating protein GYP7 [Gossypium arboreum] |
| 3 |
Hb_001801_020 |
0.0666847945 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 4 |
Hb_022250_010 |
0.0810685216 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 5 |
Hb_000359_190 |
0.0888384901 |
- |
- |
PREDICTED: protein VACUOLELESS1 [Nicotiana sylvestris] |
| 6 |
Hb_002400_060 |
0.0896258746 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 7 |
Hb_001246_130 |
0.0947450864 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000510_150 |
0.0963449595 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000562_050 |
0.097955242 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 10 |
Hb_001226_110 |
0.0983095685 |
- |
- |
PREDICTED: uncharacterized protein LOC105638106 [Jatropha curcas] |
| 11 |
Hb_000085_260 |
0.0990539451 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 12 |
Hb_021977_010 |
0.0990799076 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 13 |
Hb_002301_030 |
0.0996349578 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 14 |
Hb_008014_040 |
0.10022165 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 15 |
Hb_001541_150 |
0.1010839998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000265_140 |
0.1028188232 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 17 |
Hb_015884_020 |
0.1049455367 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 18 |
Hb_000922_060 |
0.1050499613 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 19 |
Hb_009476_120 |
0.1067991549 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 20 |
Hb_002311_130 |
0.1071754857 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |