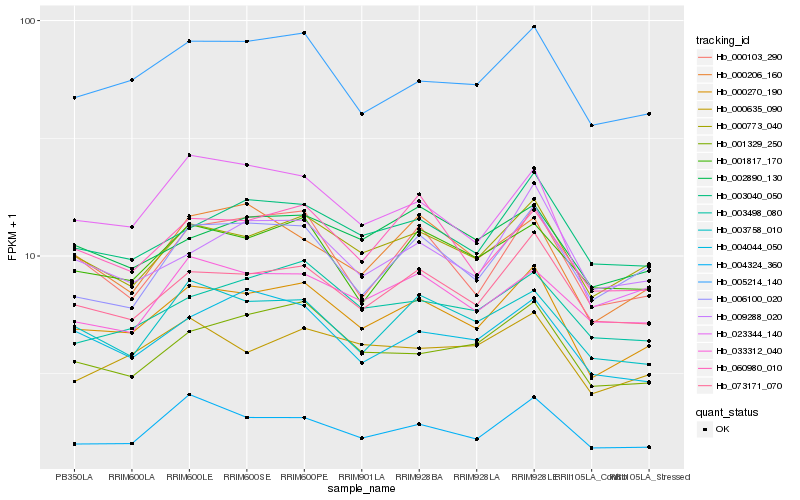
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_190 |
0.0 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 2 |
Hb_000773_040 |
0.0490262684 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 3 |
Hb_073171_070 |
0.0537395086 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 4 |
Hb_005214_140 |
0.0573189908 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 5 |
Hb_003040_050 |
0.0669957251 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 6 |
Hb_004324_360 |
0.067351936 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001329_250 |
0.069193456 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000103_290 |
0.0699536261 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 9 |
Hb_060980_010 |
0.0714980776 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 10 |
Hb_003758_010 |
0.0717945261 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001817_170 |
0.0719303622 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 12 |
Hb_006100_020 |
0.0726684811 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 13 |
Hb_009288_020 |
0.073639544 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 14 |
Hb_000635_090 |
0.0738431494 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 15 |
Hb_002890_130 |
0.0739172632 |
- |
- |
tip120, putative [Ricinus communis] |
| 16 |
Hb_023344_140 |
0.0746060088 |
- |
- |
PREDICTED: WD repeat-containing protein 70 [Jatropha curcas] |
| 17 |
Hb_033312_040 |
0.0751047956 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_003498_080 |
0.0752312437 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 19 |
Hb_000206_160 |
0.0756165731 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_004044_050 |
0.075872351 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |