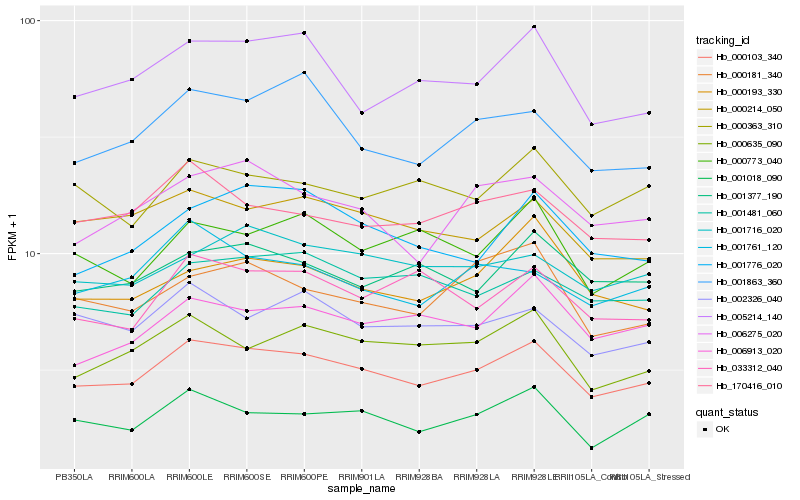
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_340 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 2 |
Hb_000214_050 |
0.0680426892 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000635_090 |
0.0680768955 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 4 |
Hb_001018_090 |
0.0683045598 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 5 |
Hb_005214_140 |
0.0684299974 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 6 |
Hb_006275_020 |
0.069038926 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 7 |
Hb_170416_010 |
0.0699431554 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 8 |
Hb_001481_060 |
0.0716327229 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001716_020 |
0.0716813984 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 10 |
Hb_006913_020 |
0.071684029 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002326_040 |
0.0725143454 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 12 |
Hb_001776_020 |
0.07309022 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 13 |
Hb_001377_190 |
0.0731083953 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001761_120 |
0.0738166927 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 15 |
Hb_001863_360 |
0.0743749449 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 16 |
Hb_000363_310 |
0.075659433 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 17 |
Hb_033312_040 |
0.0758123158 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000773_040 |
0.0759041669 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000193_330 |
0.0767782573 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 20 |
Hb_000181_340 |
0.0767818778 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |