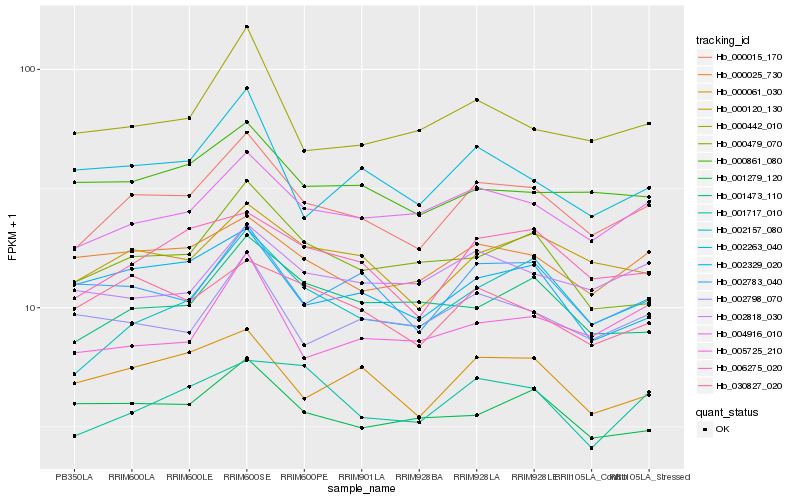
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000015_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000120_130 |
0.0636219887 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 3 |
Hb_004916_010 |
0.0644974281 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 4 |
Hb_006275_020 |
0.0658788912 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 5 |
Hb_002157_080 |
0.0738629663 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000061_030 |
0.0802154181 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065 [Jatropha curcas] |
| 7 |
Hb_000025_730 |
0.0802190569 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 8 |
Hb_002783_040 |
0.0807963444 |
transcription factor |
TF Family: IWS1 |
- |
| 9 |
Hb_001473_110 |
0.0838027703 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 10 |
Hb_000861_080 |
0.084145758 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 11 |
Hb_030827_020 |
0.0846475254 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 12 |
Hb_000442_010 |
0.0853645116 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 13 |
Hb_002329_020 |
0.0855318566 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 14 |
Hb_000479_070 |
0.0858115898 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 15 |
Hb_002818_030 |
0.0866713393 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 16 |
Hb_001717_010 |
0.0879841359 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 17 |
Hb_002798_070 |
0.0885533362 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 18 |
Hb_005725_210 |
0.0891789029 |
- |
- |
PREDICTED: RNA-binding protein 1 [Jatropha curcas] |
| 19 |
Hb_001279_120 |
0.0908801722 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 20 |
Hb_002263_040 |
0.0912497858 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |