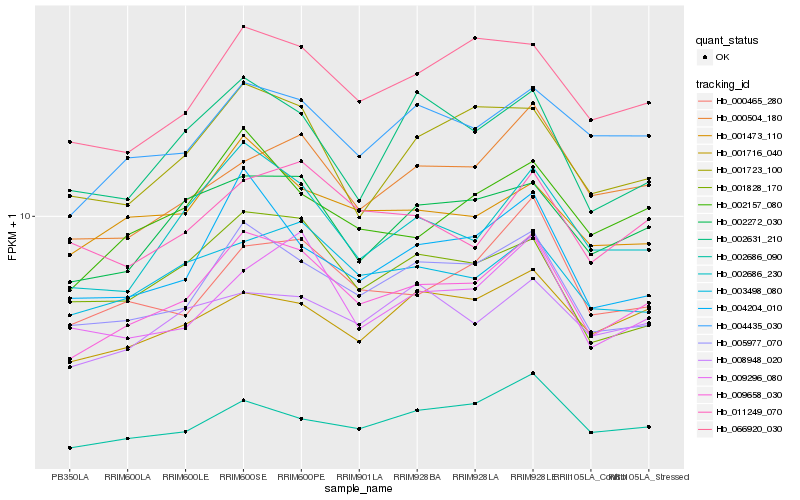
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009658_030 |
0.0 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 2 |
Hb_005977_070 |
0.063480405 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002686_230 |
0.0721901069 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_002272_030 |
0.0747927548 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 5 |
Hb_066920_030 |
0.07560397 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 6 |
Hb_004435_030 |
0.0762448802 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 7 |
Hb_002157_080 |
0.0767071131 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 8 |
Hb_004204_010 |
0.0810792252 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 9 |
Hb_003498_080 |
0.0817605911 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 10 |
Hb_000465_280 |
0.082684084 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 11 |
Hb_001723_100 |
0.085278371 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000504_180 |
0.0862050401 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 13 |
Hb_001473_110 |
0.0867457583 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 14 |
Hb_001828_170 |
0.0873143632 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 15 |
Hb_009296_080 |
0.0877231384 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002686_090 |
0.0884308794 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 17 |
Hb_001716_040 |
0.0893629213 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 18 |
Hb_008948_020 |
0.0904873844 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 19 |
Hb_011249_070 |
0.0914387519 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 20 |
Hb_002631_210 |
0.0914527149 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |