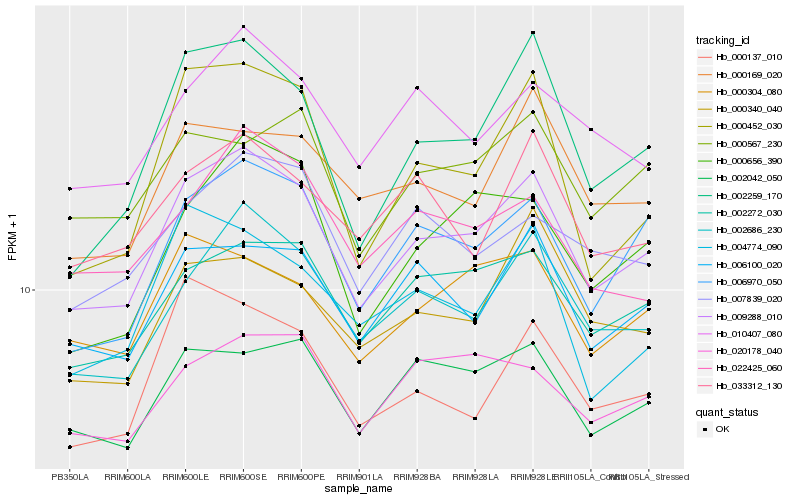
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009288_010 |
0.0 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_002686_230 |
0.062206291 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_006970_050 |
0.0652770567 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 4 |
Hb_002272_030 |
0.0703407372 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 5 |
Hb_002259_170 |
0.0722851635 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_006100_020 |
0.0741275006 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 7 |
Hb_002042_050 |
0.0783284287 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000304_080 |
0.0810361334 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_010407_080 |
0.0827539424 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000656_390 |
0.0835159125 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000169_020 |
0.0854085169 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 12 |
Hb_007839_020 |
0.0855892491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000137_010 |
0.0871139925 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 14 |
Hb_000567_230 |
0.0874035194 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 15 |
Hb_022425_060 |
0.0879698071 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 16 |
Hb_000452_030 |
0.0879988755 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 17 |
Hb_033312_130 |
0.0880606068 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000340_040 |
0.0893041323 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 19 |
Hb_020178_040 |
0.0897915831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004774_090 |
0.0905911741 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |