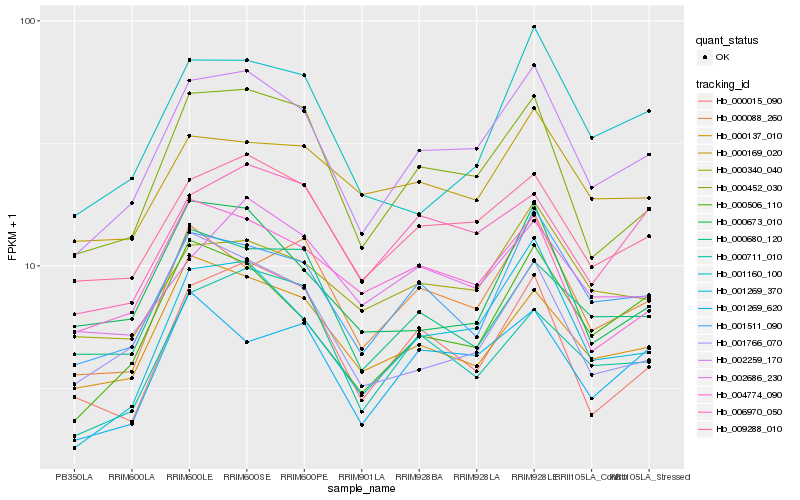
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_170 |
0.0 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_009288_010 |
0.0722851635 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_000452_030 |
0.0741250668 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 4 |
Hb_001269_370 |
0.0744619272 |
- |
- |
PREDICTED: U-box domain-containing protein 43 [Jatropha curcas] |
| 5 |
Hb_001511_090 |
0.0790026732 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 6 |
Hb_000506_110 |
0.0894465502 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Jatropha curcas] |
| 7 |
Hb_000137_010 |
0.0911990433 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 8 |
Hb_000088_260 |
0.0917369226 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 9 |
Hb_000340_040 |
0.0972725897 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 10 |
Hb_006970_050 |
0.1005847835 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 11 |
Hb_000015_090 |
0.1009472702 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 12 |
Hb_001269_620 |
0.1013227332 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 13 |
Hb_000711_010 |
0.101679647 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 14 |
Hb_004774_090 |
0.1026288823 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 15 |
Hb_000680_120 |
0.102850786 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 16 |
Hb_002686_230 |
0.105521685 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_000673_010 |
0.1062771044 |
- |
- |
PREDICTED: uncharacterized protein LOC105636554 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000169_020 |
0.1067791791 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 19 |
Hb_001766_070 |
0.1075295282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001160_100 |
0.1078855313 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 6, chloroplastic [Jatropha curcas] |