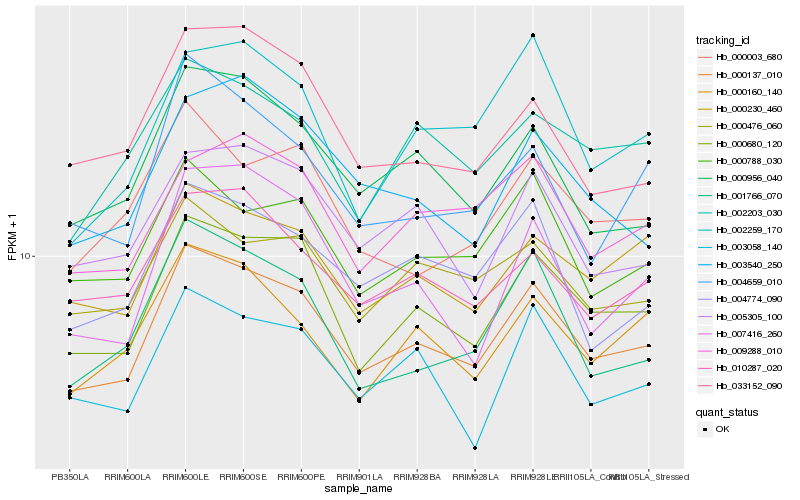
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000137_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 2 |
Hb_000680_120 |
0.0485169341 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 3 |
Hb_000956_040 |
0.0754669479 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 4 |
Hb_000230_460 |
0.0778991534 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 5 |
Hb_000788_030 |
0.0781574243 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 6 |
Hb_005305_100 |
0.0816122355 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004774_090 |
0.082344134 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 8 |
Hb_001766_070 |
0.0828284194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010287_020 |
0.0830104015 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 10 |
Hb_000476_060 |
0.0852608173 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_000160_140 |
0.0852651414 |
- |
- |
PREDICTED: ELL-associated factor 2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_003058_140 |
0.0862325147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_009288_010 |
0.0871139925 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_002203_030 |
0.0892021435 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 15 |
Hb_004659_010 |
0.0909271916 |
- |
- |
Guanine nucleotide-binding subunit beta-2 [Gossypium arboreum] |
| 16 |
Hb_000003_680 |
0.0910866942 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 17 |
Hb_033152_090 |
0.0911400147 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 18 |
Hb_003540_250 |
0.0911698841 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002259_170 |
0.0911990433 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_007416_260 |
0.0918460285 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF12-A-like [Jatropha curcas] |