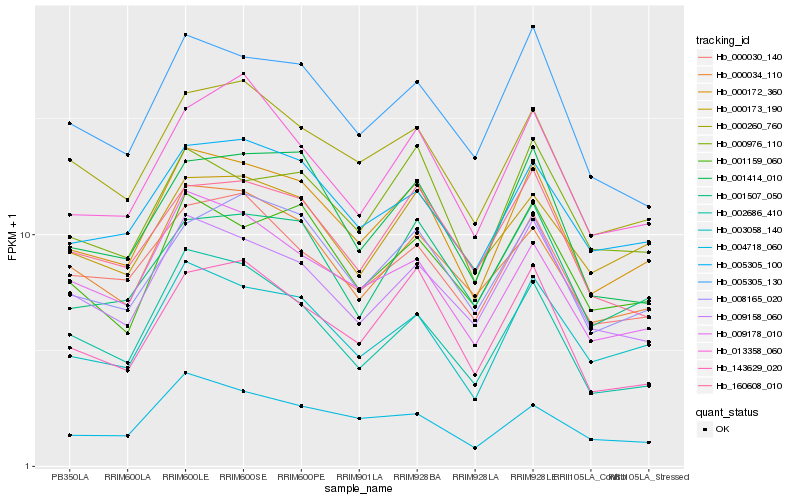
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_360 |
0.0 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 2 |
Hb_003058_140 |
0.0577527578 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005305_100 |
0.0643167233 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000260_760 |
0.0653257595 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 5 |
Hb_008165_020 |
0.0698976495 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009158_060 |
0.072865637 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 7 |
Hb_001159_060 |
0.0742299127 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 8 |
Hb_001507_050 |
0.0748104979 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000034_110 |
0.0751454005 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 10 |
Hb_009178_010 |
0.0752982525 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 11 |
Hb_001414_010 |
0.0754907465 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 12 |
Hb_000030_140 |
0.0760397517 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005305_130 |
0.0765719983 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 14 |
Hb_000976_110 |
0.077504052 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 15 |
Hb_143629_020 |
0.0792282645 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 16 |
Hb_002686_410 |
0.0816477762 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000173_190 |
0.0821676389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004718_060 |
0.0821705586 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_160608_010 |
0.0822519345 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 20 |
Hb_013358_060 |
0.0835992056 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |