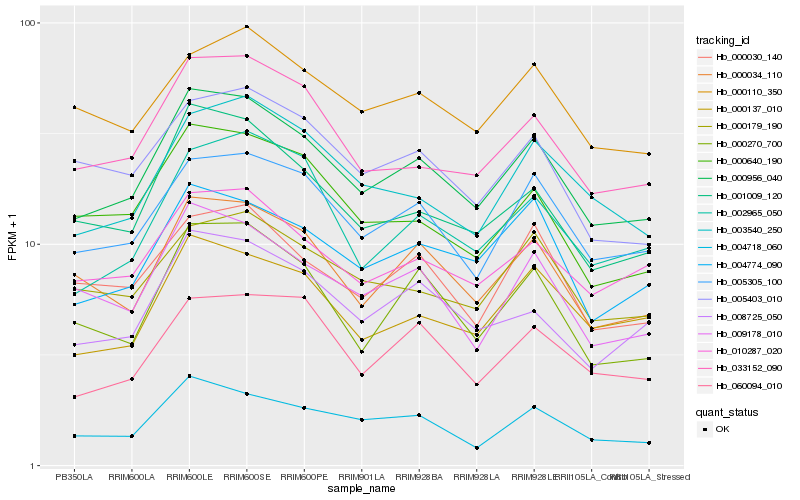
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000956_040 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 2 |
Hb_005305_100 |
0.0615437489 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000034_110 |
0.0651490856 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 4 |
Hb_033152_090 |
0.0672774382 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 5 |
Hb_001009_120 |
0.0675234477 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 6 |
Hb_010287_020 |
0.0714825441 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 7 |
Hb_004774_090 |
0.0728891719 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 8 |
Hb_009178_010 |
0.0737706142 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 9 |
Hb_008725_050 |
0.0746028469 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 10 |
Hb_004718_060 |
0.0754143516 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000137_010 |
0.0754669479 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 12 |
Hb_000270_700 |
0.0793945331 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 13 |
Hb_000030_140 |
0.0794724153 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003540_250 |
0.0798682566 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000640_190 |
0.0798822535 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000179_190 |
0.0813091925 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 17 |
Hb_002965_050 |
0.0825687713 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 18 |
Hb_060094_010 |
0.083089715 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005403_010 |
0.0841338735 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 20 |
Hb_000110_350 |
0.0845137314 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |