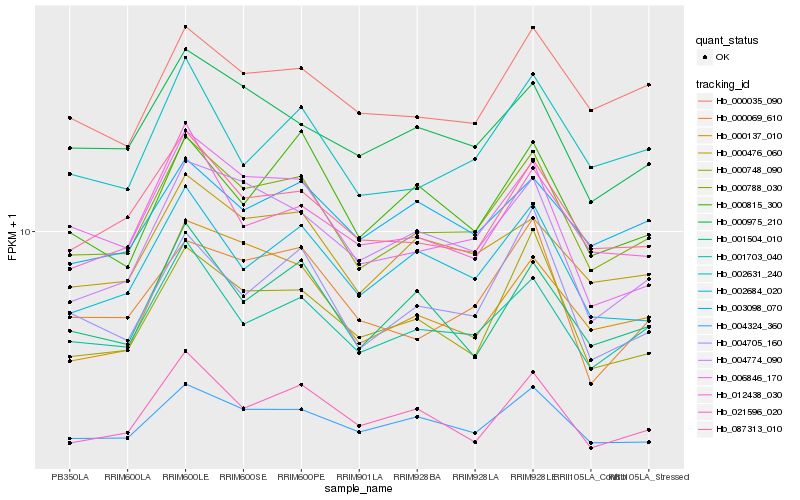
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000788_030 |
0.0 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 2 |
Hb_001703_040 |
0.0599595867 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004324_360 |
0.0614124835 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000476_060 |
0.0642145579 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_087313_010 |
0.0676814434 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002684_020 |
0.0722668568 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 7 |
Hb_006846_170 |
0.0736253778 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 8 |
Hb_000748_090 |
0.0761594655 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 9 |
Hb_002631_240 |
0.0769616419 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 10 |
Hb_004774_090 |
0.0772976412 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 11 |
Hb_000975_210 |
0.0773899539 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 12 |
Hb_004705_160 |
0.0776994728 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 13 |
Hb_000035_090 |
0.0778909643 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 14 |
Hb_001504_010 |
0.0780217477 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 15 |
Hb_000137_010 |
0.0781574243 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 16 |
Hb_021596_020 |
0.0807301814 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 17 |
Hb_012438_030 |
0.0814860623 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 18 |
Hb_000069_610 |
0.0816089219 |
- |
- |
PREDICTED: protein SUPPRESSOR OF PHYA-105 1 [Jatropha curcas] |
| 19 |
Hb_003098_070 |
0.0829709747 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 20 |
Hb_000815_300 |
0.0844580975 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |