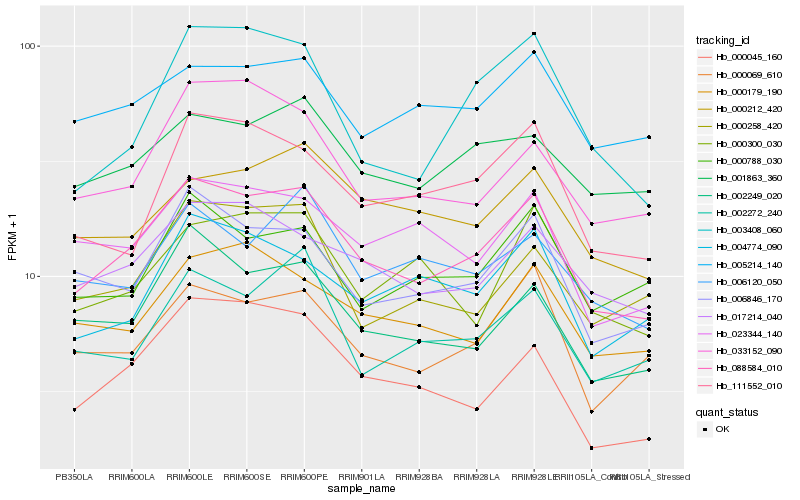
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_088584_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s08470g [Populus trichocarpa] |
| 2 |
Hb_006846_170 |
0.0822024014 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 3 |
Hb_017214_040 |
0.0840239205 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase COP1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002249_020 |
0.0860915713 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 5 |
Hb_000069_610 |
0.0913573757 |
- |
- |
PREDICTED: protein SUPPRESSOR OF PHYA-105 1 [Jatropha curcas] |
| 6 |
Hb_000179_190 |
0.0939674603 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 7 |
Hb_111552_010 |
0.0956731088 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 8 |
Hb_001863_360 |
0.0969941424 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 9 |
Hb_023344_140 |
0.0970132775 |
- |
- |
PREDICTED: WD repeat-containing protein 70 [Jatropha curcas] |
| 10 |
Hb_033152_090 |
0.0974799992 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 11 |
Hb_000045_160 |
0.0988859705 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 12 |
Hb_000212_420 |
0.0990121062 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 13 |
Hb_004774_090 |
0.0995170371 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 14 |
Hb_002272_240 |
0.099914643 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_005214_140 |
0.1012282808 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 16 |
Hb_000258_420 |
0.1018574846 |
- |
- |
sucrose transporter 2A [Hevea brasiliensis] |
| 17 |
Hb_000788_030 |
0.1018912866 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 18 |
Hb_000300_030 |
0.1039675325 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 19 |
Hb_006120_050 |
0.1044592554 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 20 |
Hb_003408_060 |
0.1047475981 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |