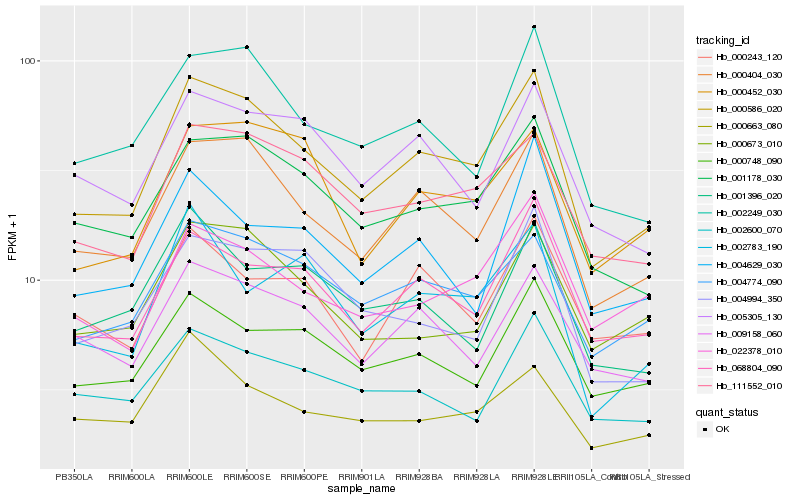
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000586_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein F383_02497 [Gossypium arboreum] |
| 2 |
Hb_000404_030 |
0.0559822674 |
- |
- |
PREDICTED: protein NAP1 [Jatropha curcas] |
| 3 |
Hb_001178_030 |
0.0786158578 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |
| 4 |
Hb_004774_090 |
0.0850842094 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 5 |
Hb_111552_010 |
0.0863484263 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 6 |
Hb_002249_030 |
0.088337101 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 7 |
Hb_068804_090 |
0.0884728389 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 8 |
Hb_000748_090 |
0.0992535255 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 9 |
Hb_009158_060 |
0.1014122528 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 10 |
Hb_005305_130 |
0.1036013536 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 11 |
Hb_000452_030 |
0.1046644872 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 12 |
Hb_000673_010 |
0.1051221861 |
- |
- |
PREDICTED: uncharacterized protein LOC105636554 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002600_070 |
0.10902693 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 14 |
Hb_004994_350 |
0.1091722042 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 15 |
Hb_022378_010 |
0.1094110533 |
- |
- |
PREDICTED: uncharacterized protein LOC105634715 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001396_020 |
0.1122432032 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 17 |
Hb_000243_120 |
0.1142809354 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_002783_190 |
0.1143740591 |
- |
- |
PREDICTED: uncharacterized protein LOC105635339 [Jatropha curcas] |
| 19 |
Hb_004629_030 |
0.1145628399 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_000663_080 |
0.114598871 |
transcription factor |
TF Family: HB |
Homeobox protein HAT3.1, putative [Ricinus communis] |