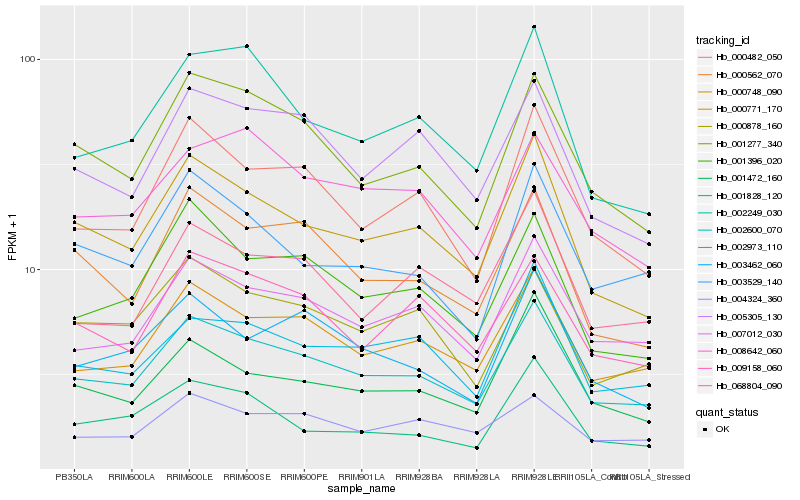
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002600_070 |
0.0 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 2 |
Hb_000748_090 |
0.051511224 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 3 |
Hb_007012_030 |
0.057498312 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000771_170 |
0.0610884907 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 5 |
Hb_001277_340 |
0.0711752829 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 6 |
Hb_000482_050 |
0.0733781307 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_009158_060 |
0.0777995491 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 8 |
Hb_001396_020 |
0.0778663784 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 9 |
Hb_005305_130 |
0.0780624567 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 10 |
Hb_000878_160 |
0.0812778214 |
- |
- |
PREDICTED: D-lactate dehydrogenase [cytochrome], mitochondrial [Jatropha curcas] |
| 11 |
Hb_000562_070 |
0.0821820165 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002973_110 |
0.0822880496 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_068804_090 |
0.0857548029 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 14 |
Hb_003462_060 |
0.0861399014 |
- |
- |
PREDICTED: ABC transporter G family member 24-like [Jatropha curcas] |
| 15 |
Hb_008642_060 |
0.0866000161 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 16 |
Hb_003529_140 |
0.086693412 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002249_030 |
0.0875717596 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004324_360 |
0.0876921361 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001828_120 |
0.0888154907 |
- |
- |
PREDICTED: uncharacterized protein LOC105140167 [Populus euphratica] |
| 20 |
Hb_001472_160 |
0.0896170426 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |