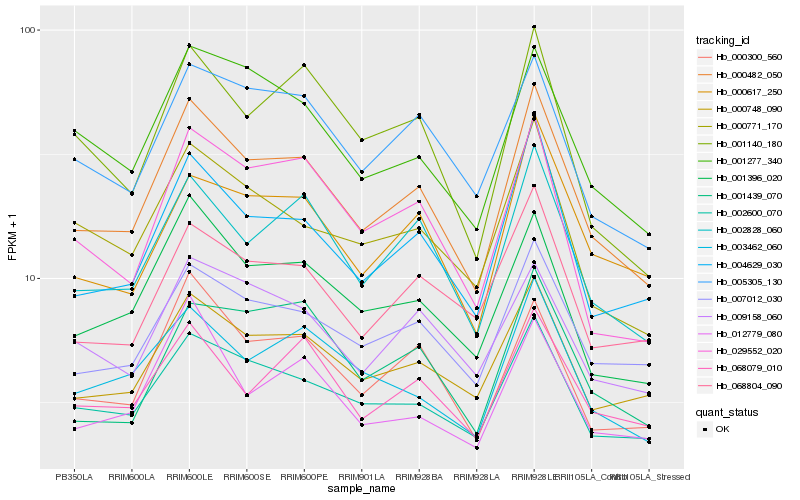
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000482_050 |
0.0 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002828_060 |
0.0648857444 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004629_030 |
0.0652748175 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_001396_020 |
0.0720421768 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 5 |
Hb_002600_070 |
0.0733781307 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 6 |
Hb_007012_030 |
0.0738203809 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000748_090 |
0.0756506496 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 8 |
Hb_000300_560 |
0.0762263839 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000617_250 |
0.0838437288 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005305_130 |
0.0849488631 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 11 |
Hb_000771_170 |
0.0849696588 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_001277_340 |
0.0851877913 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 13 |
Hb_003462_060 |
0.0859975094 |
- |
- |
PREDICTED: ABC transporter G family member 24-like [Jatropha curcas] |
| 14 |
Hb_068079_010 |
0.0862018951 |
- |
- |
- |
| 15 |
Hb_068804_090 |
0.0873060202 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 16 |
Hb_029552_020 |
0.0892440994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_012779_080 |
0.0908167899 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 18 |
Hb_009158_060 |
0.0914464067 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 19 |
Hb_001140_180 |
0.0935703905 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 20 |
Hb_001439_070 |
0.0943313991 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |