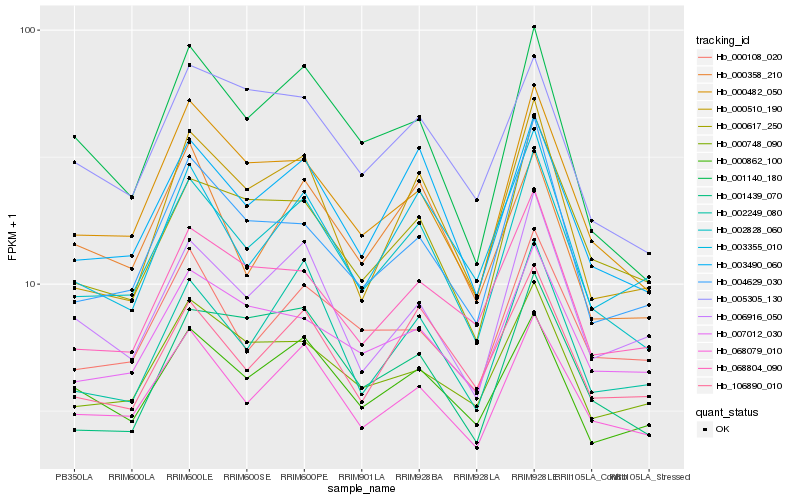
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002828_060 |
0.0 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003490_060 |
0.0539165319 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 3 |
Hb_068079_010 |
0.0633482076 |
- |
- |
- |
| 4 |
Hb_000482_050 |
0.0648857444 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000510_190 |
0.0794156276 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 6 |
Hb_002249_080 |
0.0795488026 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 7 |
Hb_001140_180 |
0.0806527241 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 8 |
Hb_000862_100 |
0.0833085212 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_106890_010 |
0.0859772461 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_004629_030 |
0.0874336828 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_006916_050 |
0.0886418249 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 12 |
Hb_005305_130 |
0.0888406625 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 13 |
Hb_000617_250 |
0.0895612489 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003355_010 |
0.0900256922 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 15 |
Hb_000748_090 |
0.091675337 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 16 |
Hb_001439_070 |
0.0921943163 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 17 |
Hb_000358_210 |
0.0923672011 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 18 |
Hb_068804_090 |
0.0924880116 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 19 |
Hb_000108_020 |
0.0926055558 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 20 |
Hb_007012_030 |
0.0928755802 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |