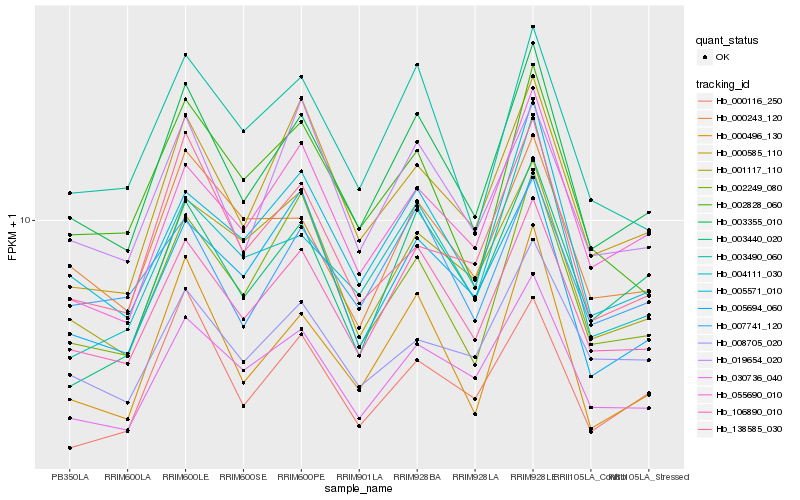
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003355_010 |
0.0 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 2 |
Hb_106890_010 |
0.0394223226 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_030736_040 |
0.0662630698 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005571_010 |
0.0686479479 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 5 |
Hb_000496_130 |
0.0747614421 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 6 |
Hb_007741_120 |
0.078452257 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003490_060 |
0.0803870178 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 8 |
Hb_055690_010 |
0.0805720308 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000585_110 |
0.080979455 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 10 |
Hb_000243_120 |
0.0843727729 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_019654_020 |
0.0844365264 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 12 |
Hb_002249_080 |
0.0851710178 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 13 |
Hb_003440_020 |
0.0864379781 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_138585_030 |
0.0865576062 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 15 |
Hb_004111_030 |
0.0884629278 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_001117_110 |
0.0887026945 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 17 |
Hb_008705_020 |
0.0895545774 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_000116_250 |
0.0897888547 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 19 |
Hb_002828_060 |
0.0900256922 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_005694_060 |
0.0914491042 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |