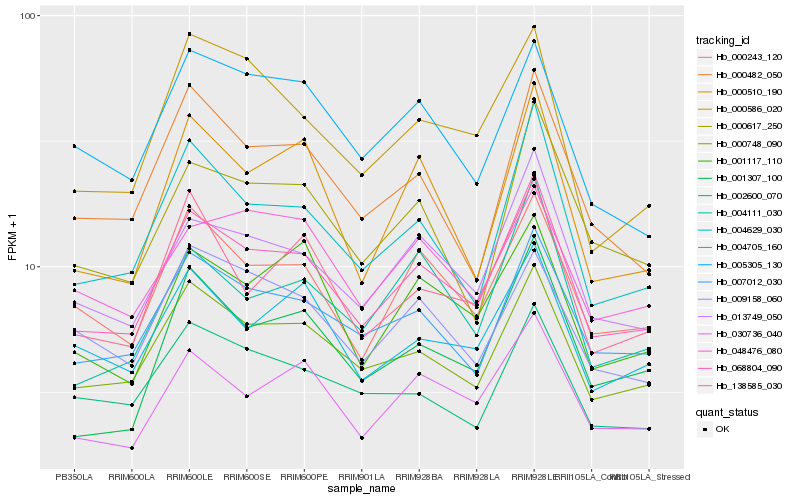
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_068804_090 |
0.0 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 2 |
Hb_000748_090 |
0.053820734 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 3 |
Hb_013749_050 |
0.0541082933 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 4 |
Hb_007012_030 |
0.0620306839 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000243_120 |
0.0642320929 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_030736_040 |
0.0669795377 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004629_030 |
0.0679907006 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_004705_160 |
0.081473867 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 9 |
Hb_001117_110 |
0.0816160303 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 10 |
Hb_138585_030 |
0.0832520874 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 11 |
Hb_000617_250 |
0.0839230143 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002600_070 |
0.0857548029 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 13 |
Hb_001307_100 |
0.0859997357 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 14 |
Hb_000482_050 |
0.0873060202 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000586_020 |
0.0884728389 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein F383_02497 [Gossypium arboreum] |
| 16 |
Hb_000510_190 |
0.0894294433 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 17 |
Hb_004111_030 |
0.0898166773 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_009158_060 |
0.0902850007 |
- |
- |
PREDICTED: CDPK-related kinase 3 [Jatropha curcas] |
| 19 |
Hb_005305_130 |
0.0905687492 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 20 |
Hb_048476_080 |
0.0922115768 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |