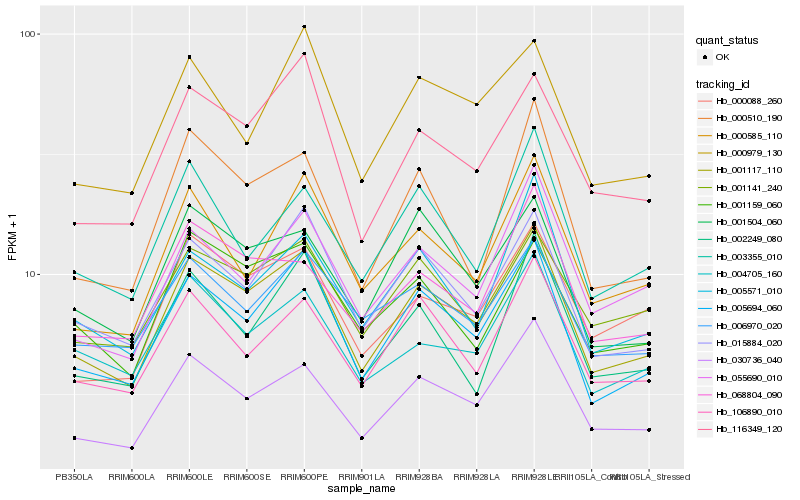
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001117_110 |
0.0 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 2 |
Hb_005694_060 |
0.0470459853 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 3 |
Hb_030736_040 |
0.0527684712 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 4 |
Hb_015884_020 |
0.0673645143 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 5 |
Hb_116349_120 |
0.0741477371 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_106890_010 |
0.0771263839 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_000088_260 |
0.0785249275 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 8 |
Hb_001141_240 |
0.0799479497 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001504_060 |
0.079981316 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 10 |
Hb_055690_010 |
0.0802895559 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_068804_090 |
0.0816160303 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 12 |
Hb_004705_160 |
0.0820467625 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 13 |
Hb_000585_110 |
0.0835549177 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 14 |
Hb_005571_010 |
0.084983123 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 15 |
Hb_001159_060 |
0.0855069347 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 16 |
Hb_006970_020 |
0.0859686551 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 17 |
Hb_000979_130 |
0.0861078845 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 18 |
Hb_002249_080 |
0.0880939319 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 19 |
Hb_000510_190 |
0.0882631028 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 20 |
Hb_003355_010 |
0.0887026945 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |