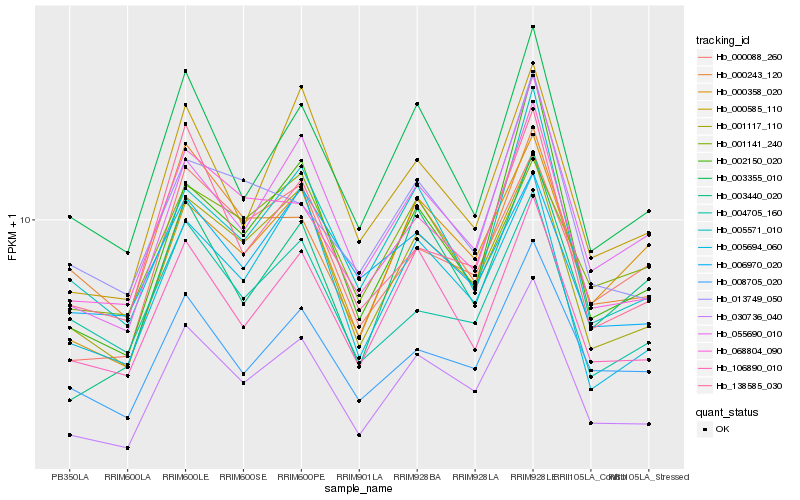
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030736_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001117_110 |
0.0527684712 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 3 |
Hb_106890_010 |
0.0562264297 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_055690_010 |
0.061360663 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003355_010 |
0.0662630698 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 6 |
Hb_068804_090 |
0.0669795377 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 7 |
Hb_008705_020 |
0.0682303033 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_000358_020 |
0.0746127489 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 9 |
Hb_005571_010 |
0.0758793159 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 10 |
Hb_000585_110 |
0.0785874915 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 11 |
Hb_005694_060 |
0.0789747306 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 12 |
Hb_000243_120 |
0.0796979438 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000088_260 |
0.0823980648 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 14 |
Hb_004705_160 |
0.0831897924 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 15 |
Hb_013749_050 |
0.0833132183 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 16 |
Hb_138585_030 |
0.0842430146 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 17 |
Hb_002150_020 |
0.0850902165 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_006970_020 |
0.0858063425 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 19 |
Hb_001141_240 |
0.0862214118 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003440_020 |
0.0863015147 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |