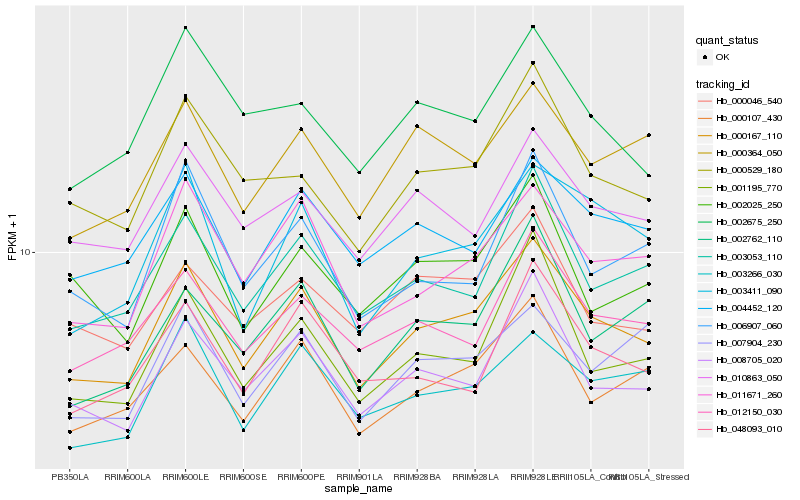
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000167_110 |
0.0 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_011671_260 |
0.0675589732 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008705_020 |
0.0708126626 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_000107_430 |
0.0749329956 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 5 |
Hb_003053_110 |
0.0749647106 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003411_090 |
0.0753311616 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_003266_030 |
0.076771705 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 8 |
Hb_004452_120 |
0.0778873721 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 9 |
Hb_012150_030 |
0.079089317 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000529_180 |
0.0794961981 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001195_770 |
0.0812161188 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 12 |
Hb_000364_050 |
0.0824464657 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_010863_050 |
0.0827861168 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 14 |
Hb_006907_060 |
0.0869039494 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_002025_250 |
0.0876436911 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 16 |
Hb_000046_540 |
0.0877352063 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 17 |
Hb_048093_010 |
0.0886438258 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_002675_250 |
0.0888175449 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 19 |
Hb_002762_110 |
0.0889961672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007904_230 |
0.0899998184 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |