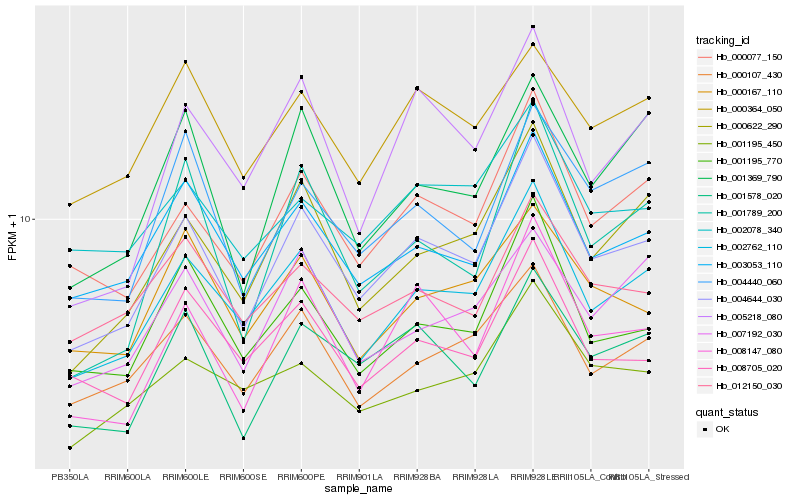
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004644_030 |
0.0 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 2 |
Hb_002762_110 |
0.0548459788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000077_150 |
0.0611242984 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |
| 4 |
Hb_000622_290 |
0.0714811152 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 5 |
Hb_008147_080 |
0.0730987724 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003053_110 |
0.074238341 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001578_020 |
0.0776624038 |
- |
- |
Protein virR, putative [Ricinus communis] |
| 8 |
Hb_001195_770 |
0.079621748 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 9 |
Hb_000107_430 |
0.0810234523 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 10 |
Hb_004440_060 |
0.0868663521 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 11 |
Hb_008705_020 |
0.0893829753 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_001195_450 |
0.0899051952 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 13 |
Hb_012150_030 |
0.0906848034 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000364_050 |
0.0908109427 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001369_790 |
0.0914928607 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000167_110 |
0.0919084933 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_002078_340 |
0.0919296652 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 18 |
Hb_007192_030 |
0.0928266972 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 19 |
Hb_001789_200 |
0.0932078577 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 20 |
Hb_005218_080 |
0.0947984941 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |