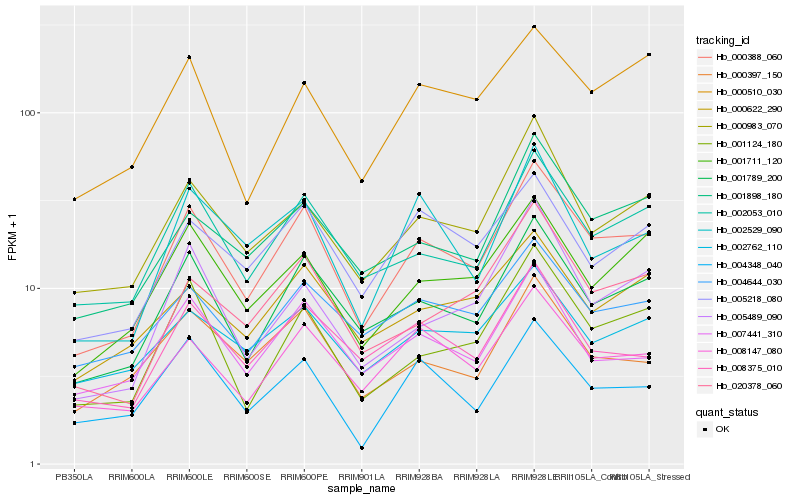
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000388_060 |
0.0 |
- |
- |
fructokinase [Manihot esculenta] |
| 2 |
Hb_001789_200 |
0.0737747194 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002053_010 |
0.0901154309 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_008147_080 |
0.093853008 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000983_070 |
0.097571688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001124_180 |
0.099715991 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 7 |
Hb_001898_180 |
0.1027548928 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001711_120 |
0.1041776465 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_020378_060 |
0.1072519153 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 10 |
Hb_008375_010 |
0.1077264356 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 11 |
Hb_004644_030 |
0.1100730034 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 12 |
Hb_005218_080 |
0.1100869267 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_002529_090 |
0.1114376157 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 14 |
Hb_007441_310 |
0.1119646995 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000510_030 |
0.1134845743 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 16 |
Hb_000397_150 |
0.1148630864 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 17 |
Hb_004348_040 |
0.1153406899 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 18 |
Hb_000622_290 |
0.1162434583 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 19 |
Hb_005489_090 |
0.1169215786 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 20 |
Hb_002762_110 |
0.11708138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |