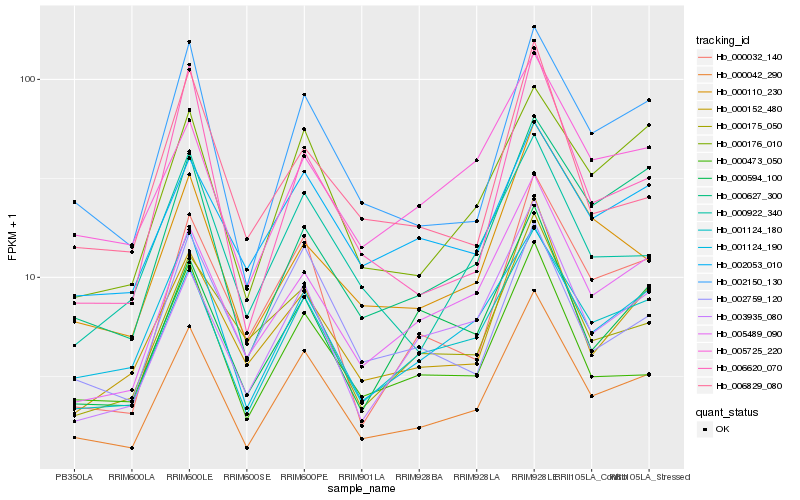
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_290 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001124_180 |
0.0842240824 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 3 |
Hb_002150_130 |
0.0911524318 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_005489_090 |
0.1028295284 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000152_480 |
0.1064272278 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003935_080 |
0.1086521539 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000176_010 |
0.1096475998 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000032_140 |
0.1100380744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001124_190 |
0.1124099654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000473_050 |
0.1128832798 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 11 |
Hb_000175_050 |
0.1170314735 |
- |
- |
- |
| 12 |
Hb_000922_340 |
0.1176529336 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 13 |
Hb_000110_230 |
0.1224466976 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_006620_070 |
0.1252493458 |
- |
- |
chloroplast 30S ribosomal protein S13 [Hevea brasiliensis] |
| 15 |
Hb_000594_100 |
0.1261663812 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000627_300 |
0.1267078345 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 17 |
Hb_002759_120 |
0.1292114055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005725_220 |
0.1324841513 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 19 |
Hb_006829_080 |
0.1337689219 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 20 |
Hb_002053_010 |
0.1344501432 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |