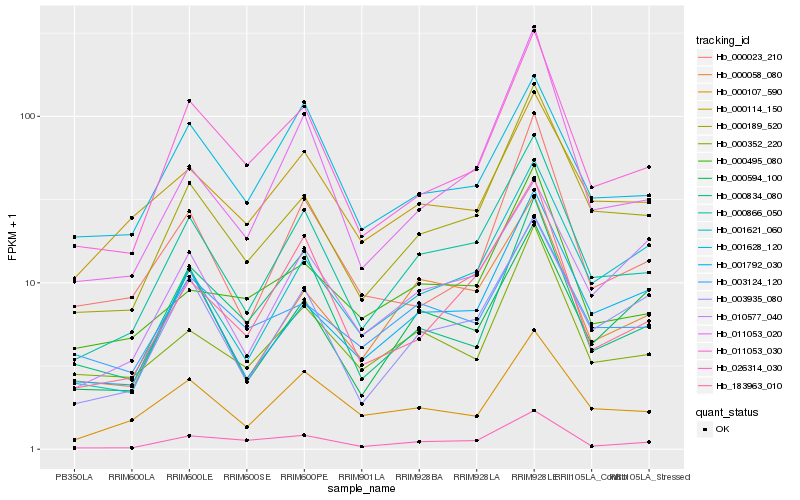
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000866_050 |
0.0 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 2 |
Hb_000058_080 |
0.0932622027 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 3 |
Hb_000189_520 |
0.0935001879 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 4 |
Hb_003935_080 |
0.1047139863 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_011053_030 |
0.1056872645 |
- |
- |
- |
| 6 |
Hb_183963_010 |
0.1072797727 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |
| 7 |
Hb_000107_590 |
0.1124932928 |
- |
- |
PREDICTED: tricalbin-3 [Jatropha curcas] |
| 8 |
Hb_026314_030 |
0.1138629998 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 9 |
Hb_001792_030 |
0.1149137147 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 10 |
Hb_000352_220 |
0.1234710639 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_011053_020 |
0.1259154777 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 12 |
Hb_000114_150 |
0.1286419001 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 13 |
Hb_000834_080 |
0.1290894285 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 14 |
Hb_000495_080 |
0.1335744747 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 15 |
Hb_010577_040 |
0.1354288877 |
- |
- |
PREDICTED: uncharacterized protein LOC105643738 [Jatropha curcas] |
| 16 |
Hb_000023_210 |
0.1360447776 |
- |
- |
PREDICTED: protein TIC 55, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001621_060 |
0.1362456186 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003124_120 |
0.1365724315 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 19 |
Hb_000594_100 |
0.1374808192 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001628_120 |
0.1379116274 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |