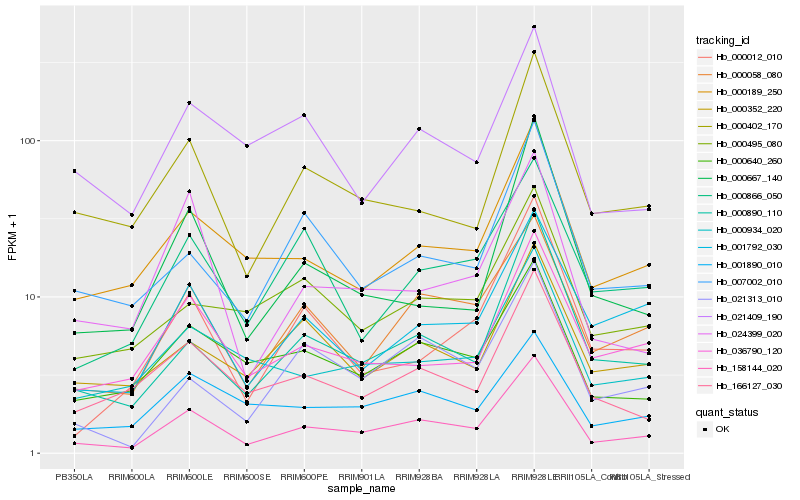
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158144_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |
| 2 |
Hb_000058_080 |
0.113963253 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 3 |
Hb_000890_110 |
0.124381527 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 4 |
Hb_000189_250 |
0.1280132123 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000640_260 |
0.1352097212 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000402_170 |
0.1372447244 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 7 |
Hb_166127_030 |
0.1394082249 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_036790_120 |
0.1409411267 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001792_030 |
0.1421341468 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 10 |
Hb_000934_020 |
0.1425933536 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |
| 11 |
Hb_021313_010 |
0.1441710473 |
- |
- |
hypothetical protein POPTR_0010s24380g [Populus trichocarpa] |
| 12 |
Hb_007002_010 |
0.146361292 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 13 |
Hb_000352_220 |
0.1499697523 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000866_050 |
0.1530964501 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 15 |
Hb_000495_080 |
0.1550974511 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 16 |
Hb_021409_190 |
0.1551535628 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 17 |
Hb_000667_140 |
0.156956496 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_024399_020 |
0.1570525679 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 19 |
Hb_000012_010 |
0.1578963904 |
- |
- |
hypothetical protein JCGZ_07709 [Jatropha curcas] |
| 20 |
Hb_001890_010 |
0.1579636218 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |