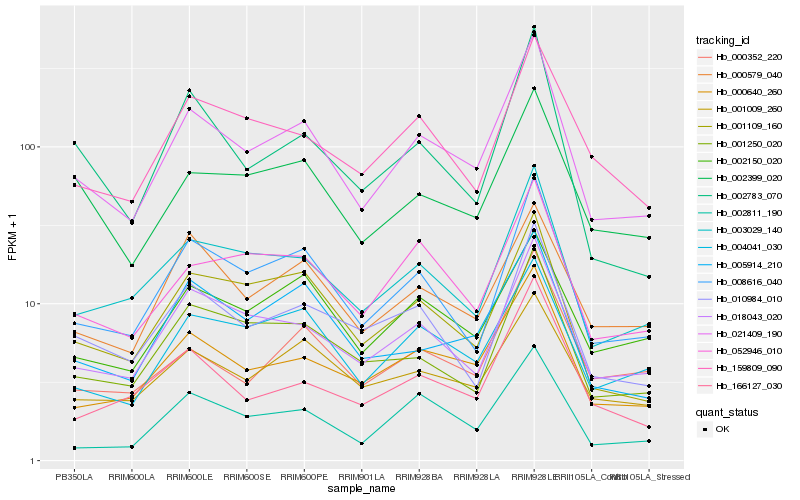
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021409_190 |
0.0 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 2 |
Hb_008616_040 |
0.0844581437 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000640_260 |
0.0853319884 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003029_140 |
0.0927101068 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 5 |
Hb_018043_020 |
0.0965310768 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001109_160 |
0.1042438932 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_002811_190 |
0.1072880488 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 8 |
Hb_004041_030 |
0.1082987735 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 9 |
Hb_005914_210 |
0.1104284498 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 10 |
Hb_052946_010 |
0.1111506955 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 11 |
Hb_001250_020 |
0.112164904 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_010984_010 |
0.113369116 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_002783_070 |
0.1137660446 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 14 |
Hb_001009_260 |
0.1151853557 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002399_020 |
0.1159874776 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 16 |
Hb_166127_030 |
0.1178702597 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000579_040 |
0.1188583247 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 18 |
Hb_000352_220 |
0.1192877701 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_159809_090 |
0.1194193136 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 20 |
Hb_002150_020 |
0.1210925496 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |