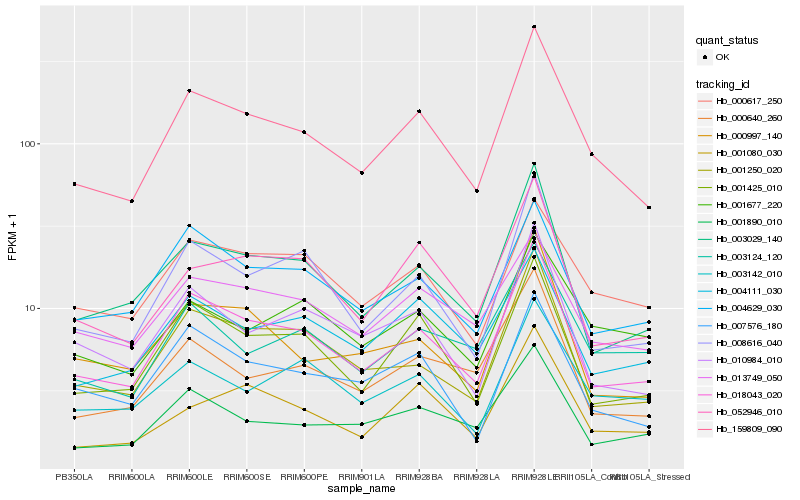
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_159809_090 |
0.0 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 2 |
Hb_018043_020 |
0.0640517181 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001425_010 |
0.0933330445 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 4 |
Hb_003029_140 |
0.1017990346 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 5 |
Hb_008616_040 |
0.1041821875 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 6 |
Hb_052946_010 |
0.1067746314 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 7 |
Hb_007576_180 |
0.1075200944 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 8 |
Hb_001890_010 |
0.1091535331 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 9 |
Hb_010984_010 |
0.1097738326 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_001250_020 |
0.1100672431 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_013749_050 |
0.1109624809 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004111_030 |
0.1110320006 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_001080_030 |
0.1114789849 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 14 |
Hb_000617_250 |
0.1124897163 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000640_260 |
0.1142348696 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004629_030 |
0.1146723558 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_003124_120 |
0.1152454915 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 18 |
Hb_000997_140 |
0.1159801597 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001677_220 |
0.1162473993 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 20 |
Hb_003142_010 |
0.1173242598 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |