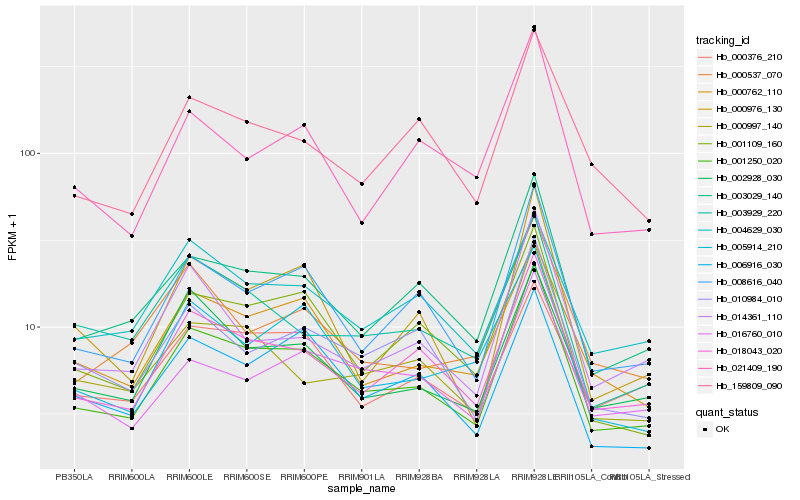
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001250_020 |
0.0 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_018043_020 |
0.0687169959 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_008616_040 |
0.0715318365 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003029_140 |
0.0755002978 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001109_160 |
0.0906074457 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000762_110 |
0.0964293423 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_010984_010 |
0.0977772006 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_000997_140 |
0.1025705607 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002928_030 |
0.102821308 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 10 |
Hb_006916_030 |
0.1075321775 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 11 |
Hb_000976_130 |
0.1080208719 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 12 |
Hb_005914_210 |
0.1085989479 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 13 |
Hb_159809_090 |
0.1100672431 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 14 |
Hb_021409_190 |
0.112164904 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 15 |
Hb_016760_010 |
0.1136178758 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_014361_110 |
0.1145068229 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_000376_210 |
0.1161654076 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 18 |
Hb_004629_030 |
0.1165533771 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_000537_070 |
0.1172749876 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 20 |
Hb_003929_220 |
0.1176797811 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |