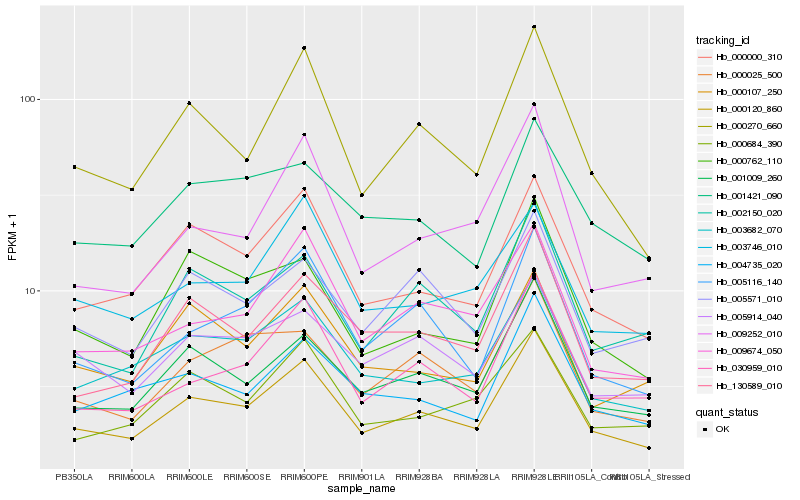
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_860 |
0.0 |
- |
- |
nucellin, putative [Ricinus communis] |
| 2 |
Hb_000270_660 |
0.0709005891 |
- |
- |
purine permease, putative [Ricinus communis] |
| 3 |
Hb_001009_260 |
0.0716910466 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000000_310 |
0.0801553845 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 5 |
Hb_030959_010 |
0.084393085 |
- |
- |
cullin, putative [Ricinus communis] |
| 6 |
Hb_005116_140 |
0.0884392492 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 7 |
Hb_003682_070 |
0.0900152 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004735_020 |
0.0900264986 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 9 |
Hb_130589_010 |
0.0909478593 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 10 |
Hb_002150_020 |
0.0931885149 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_009674_050 |
0.09667464 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 12 |
Hb_000025_500 |
0.099119763 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 13 |
Hb_009252_010 |
0.1001947768 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 14 |
Hb_000762_110 |
0.1014891287 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005571_010 |
0.1028752992 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 16 |
Hb_000684_390 |
0.1029687112 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 17 |
Hb_005914_040 |
0.1068274547 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 18 |
Hb_003746_010 |
0.1089006663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000107_250 |
0.1093471695 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 20 |
Hb_001421_090 |
0.1105670083 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |