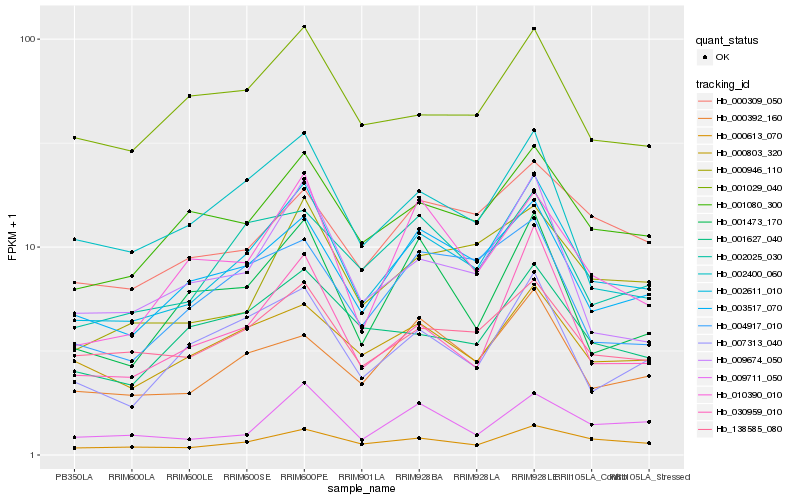
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002611_010 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_003517_070 |
0.0880553937 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 3 |
Hb_000613_070 |
0.0915517793 |
- |
- |
- |
| 4 |
Hb_001080_300 |
0.0966792428 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 5 |
Hb_138585_080 |
0.099289463 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 6 |
Hb_009674_050 |
0.1046362967 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 7 |
Hb_030959_010 |
0.104849352 |
- |
- |
cullin, putative [Ricinus communis] |
| 8 |
Hb_007313_040 |
0.1071762429 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 9 |
Hb_000803_320 |
0.1071859584 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 10 |
Hb_002025_030 |
0.1081003996 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 11 |
Hb_000946_110 |
0.1095944146 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 12 |
Hb_000392_160 |
0.1102462728 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001473_170 |
0.1139088767 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 14 |
Hb_010390_010 |
0.1139422329 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 15 |
Hb_001627_040 |
0.1148446077 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001029_040 |
0.1157839471 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 17 |
Hb_002400_060 |
0.1177228592 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 18 |
Hb_004917_010 |
0.1186365817 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 19 |
Hb_009711_050 |
0.1197219064 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 20 |
Hb_000309_050 |
0.1197294597 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |