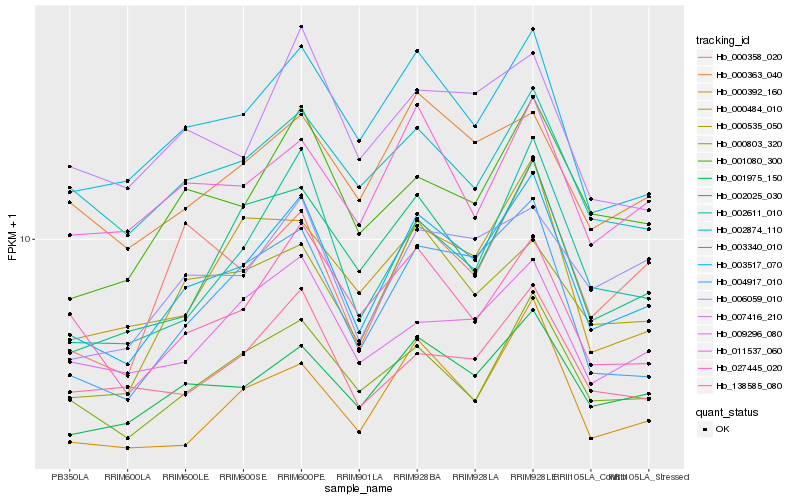
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003517_070 |
0.0 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 2 |
Hb_003340_010 |
0.0730150015 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 3 |
Hb_004917_010 |
0.0750898672 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 4 |
Hb_001975_150 |
0.0759258635 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 5 |
Hb_000392_160 |
0.0850889406 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000803_320 |
0.0855946302 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 7 |
Hb_001080_300 |
0.0861819702 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 8 |
Hb_002611_010 |
0.0880553937 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_006059_010 |
0.0942642272 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 10 |
Hb_002025_030 |
0.0949859277 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 11 |
Hb_011537_060 |
0.0950690404 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 12 |
Hb_002874_110 |
0.0950988929 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 13 |
Hb_000358_020 |
0.0955081769 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 14 |
Hb_000363_040 |
0.0956518837 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 15 |
Hb_138585_080 |
0.0966083343 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 16 |
Hb_027445_020 |
0.0966617833 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 17 |
Hb_009296_080 |
0.0978540484 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000484_010 |
0.0990579977 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 23 [Jatropha curcas] |
| 19 |
Hb_007416_210 |
0.0999337396 |
- |
- |
PREDICTED: coatomer subunit beta-1 [Jatropha curcas] |
| 20 |
Hb_000535_050 |
0.1001112527 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |