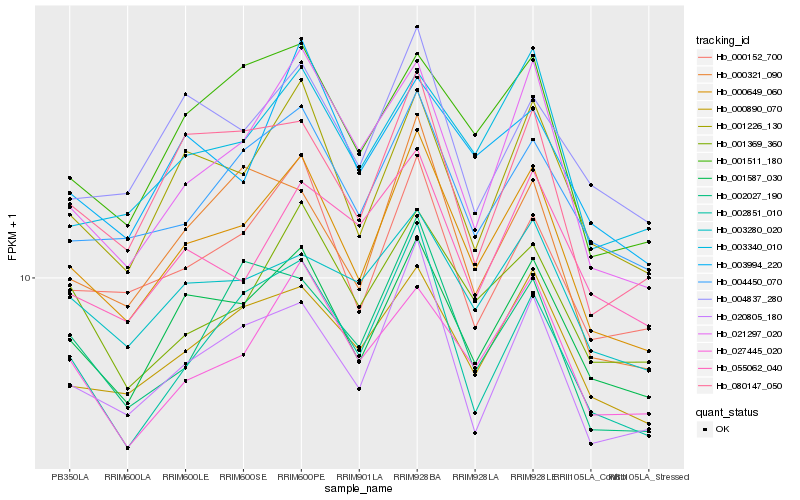
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_060 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 2 |
Hb_001587_030 |
0.0563427421 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 3 |
Hb_001226_130 |
0.0649868351 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 4 |
Hb_020805_180 |
0.069733858 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 5 |
Hb_000890_070 |
0.0720477877 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004450_070 |
0.0742551963 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 7 |
Hb_000152_700 |
0.0757613965 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |
| 8 |
Hb_021297_020 |
0.0776016556 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 9 |
Hb_003340_010 |
0.0796920771 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 10 |
Hb_002851_010 |
0.0802296672 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_001511_180 |
0.0814267088 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 12 |
Hb_027445_020 |
0.0819437487 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 13 |
Hb_001369_360 |
0.0834367282 |
- |
- |
embryo yellow protein, partial [Manihot esculenta] |
| 14 |
Hb_080147_050 |
0.086773073 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 15 |
Hb_003280_020 |
0.0892944968 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000321_090 |
0.0912694952 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 17 |
Hb_003994_220 |
0.0914530571 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 18 |
Hb_004837_280 |
0.0916388066 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 19 |
Hb_002027_190 |
0.0928386913 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 20 |
Hb_055062_040 |
0.0932731851 |
- |
- |
ceramidase, putative [Ricinus communis] |