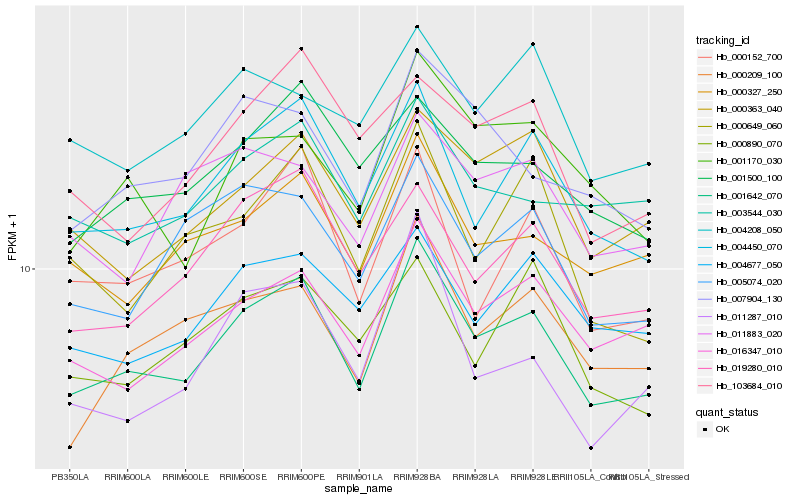
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001642_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 2 |
Hb_004450_070 |
0.0664910664 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 3 |
Hb_016347_010 |
0.0777473996 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 4 |
Hb_000327_250 |
0.0845985353 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 5 |
Hb_019280_010 |
0.0848614693 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000363_040 |
0.0867061271 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 7 |
Hb_000209_100 |
0.0868954539 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001500_100 |
0.0884262733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000152_700 |
0.0898071868 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |
| 10 |
Hb_004677_050 |
0.0905872549 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_007904_130 |
0.0909320872 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 12 |
Hb_001170_030 |
0.0919574188 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 13 |
Hb_005074_020 |
0.0924479738 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003544_030 |
0.0928969397 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 15 |
Hb_000649_060 |
0.0964893883 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 16 |
Hb_000890_070 |
0.0995461438 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004208_050 |
0.1003241234 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 18 |
Hb_103684_010 |
0.1009011284 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana tomentosiformis] |
| 19 |
Hb_011287_010 |
0.1015876147 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_011883_020 |
0.1021534453 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |