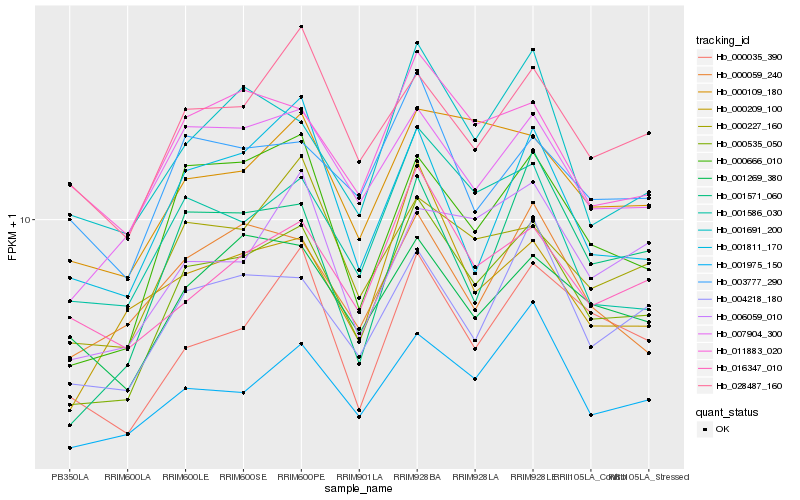
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000535_050 |
0.0 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Jatropha curcas] |
| 2 |
Hb_000666_010 |
0.06966827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007904_300 |
0.07272708 |
- |
- |
copine, putative [Ricinus communis] |
| 4 |
Hb_004218_180 |
0.0846325916 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 5 |
Hb_000227_160 |
0.085109698 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 6 |
Hb_001586_030 |
0.0878550181 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 7 |
Hb_000109_180 |
0.0881949824 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 8 |
Hb_001975_150 |
0.0894273425 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 9 |
Hb_001691_200 |
0.0898290959 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 10 |
Hb_000209_100 |
0.0933623261 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000035_390 |
0.0934631023 |
- |
- |
hypothetical protein CISIN_1g003355mg [Citrus sinensis] |
| 12 |
Hb_016347_010 |
0.0935180654 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 13 |
Hb_028487_160 |
0.0942040282 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 14 |
Hb_011883_020 |
0.0943309142 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 15 |
Hb_001571_060 |
0.095481974 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 16 |
Hb_000059_240 |
0.0956324798 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 17 |
Hb_001269_380 |
0.0958506007 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006059_010 |
0.0974127898 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 19 |
Hb_003777_290 |
0.0975767455 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 20 |
Hb_001811_170 |
0.0977978839 |
- |
- |
dynamin, putative [Ricinus communis] |