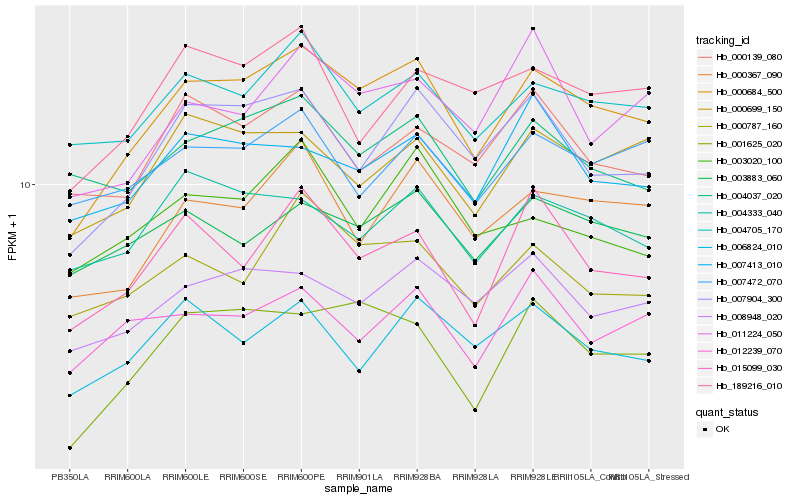
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_500 |
0.0 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 2 |
Hb_007904_300 |
0.0742552249 |
- |
- |
copine, putative [Ricinus communis] |
| 3 |
Hb_000367_090 |
0.0840494094 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 4 |
Hb_008948_020 |
0.0847024441 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 5 |
Hb_004705_170 |
0.0857438395 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 6 |
Hb_000699_150 |
0.0905350933 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 7 |
Hb_015099_030 |
0.0906772018 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 8 |
Hb_000787_160 |
0.0915062616 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004333_040 |
0.0927133964 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 10 |
Hb_003883_060 |
0.0930081932 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004037_020 |
0.0956108498 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007413_010 |
0.0962445543 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 13 |
Hb_011224_050 |
0.0968981419 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Populus euphratica] |
| 14 |
Hb_189216_010 |
0.097218575 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006824_010 |
0.0991359489 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 16 |
Hb_003020_100 |
0.0994805428 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 17 |
Hb_001625_020 |
0.0995735421 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 18 |
Hb_007472_070 |
0.1002711204 |
- |
- |
cir, putative [Ricinus communis] |
| 19 |
Hb_012239_070 |
0.1003172474 |
- |
- |
PREDICTED: uncharacterized protein LOC105634113 [Jatropha curcas] |
| 20 |
Hb_000139_080 |
0.10041343 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |