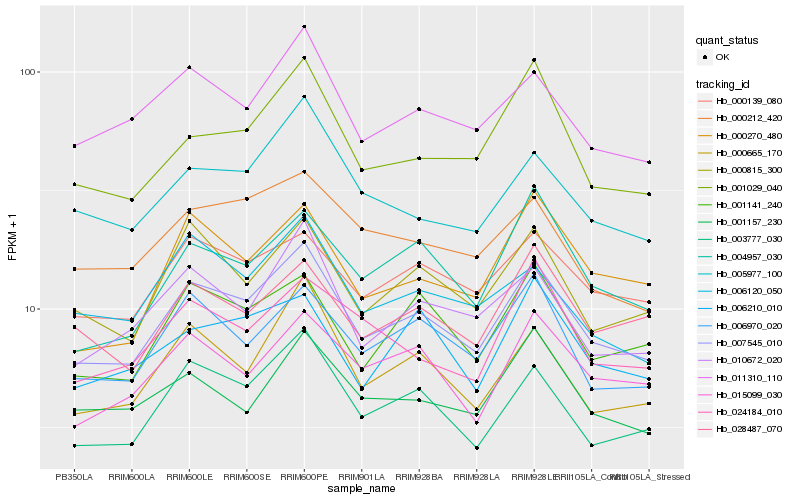
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007545_010 |
0.0 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 2 |
Hb_003777_030 |
0.0588239697 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 3 |
Hb_000815_300 |
0.0695634603 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 4 |
Hb_011310_110 |
0.0703660176 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006970_020 |
0.0712178993 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 6 |
Hb_000665_170 |
0.0716071819 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 7 |
Hb_000139_080 |
0.0719511563 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001029_040 |
0.0722108375 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 9 |
Hb_010672_020 |
0.0737106936 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_001141_240 |
0.0746444056 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005977_100 |
0.0773509044 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 12 |
Hb_015099_030 |
0.0789023219 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 13 |
Hb_004957_030 |
0.0798686086 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 14 |
Hb_028487_070 |
0.08167343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000270_480 |
0.0818801281 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 16 |
Hb_000212_420 |
0.0833834955 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 17 |
Hb_001157_230 |
0.0834180945 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 18 |
Hb_006120_050 |
0.083777757 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 19 |
Hb_024184_010 |
0.087005066 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 20 |
Hb_006210_010 |
0.0873910694 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |