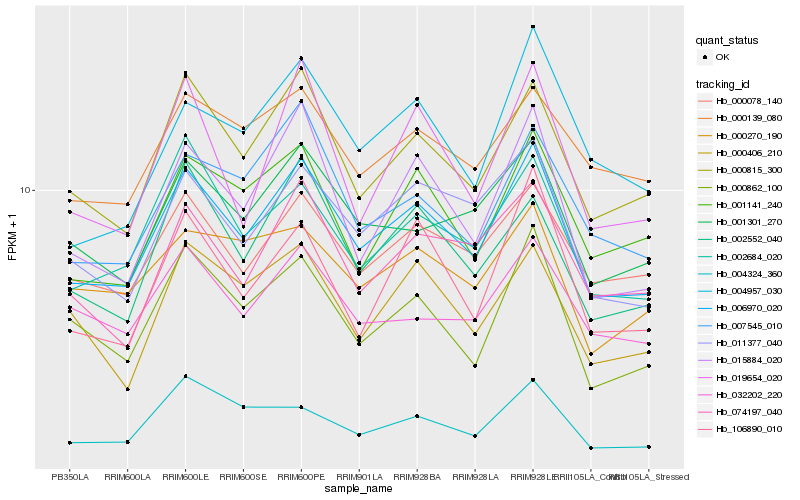
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006970_020 |
0.0 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 2 |
Hb_000815_300 |
0.0528012064 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 3 |
Hb_011377_040 |
0.0632750383 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 4 |
Hb_002684_020 |
0.0650207447 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 5 |
Hb_000078_140 |
0.0660578456 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 6 |
Hb_000862_100 |
0.0664374748 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_015884_020 |
0.0675830665 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 8 |
Hb_001301_270 |
0.0677749147 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 9 |
Hb_002552_040 |
0.0691095994 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 10 |
Hb_007545_010 |
0.0712178993 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 11 |
Hb_001141_240 |
0.071979539 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_074197_040 |
0.0721774626 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 13 |
Hb_000139_080 |
0.0786806507 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_004324_360 |
0.0790871748 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000406_210 |
0.0793299001 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 16 |
Hb_106890_010 |
0.0797118603 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_004957_030 |
0.0801652437 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 18 |
Hb_032202_220 |
0.0805710956 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_019654_020 |
0.0820621034 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 20 |
Hb_000270_190 |
0.0821853534 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |