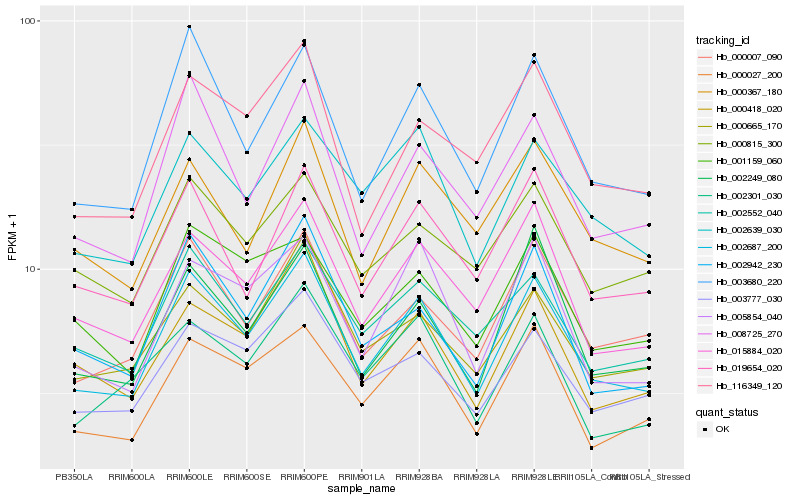
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002687_200 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003680_220 |
0.0628231285 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 3 |
Hb_015884_020 |
0.0692507019 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 4 |
Hb_000007_090 |
0.0718084069 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 5 |
Hb_000027_200 |
0.0724268983 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 6 |
Hb_002552_040 |
0.0778746537 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 7 |
Hb_002942_230 |
0.0782213614 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 8 |
Hb_003777_030 |
0.0783938612 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 9 |
Hb_008725_270 |
0.0788391762 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 10 |
Hb_000665_170 |
0.0793485149 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 11 |
Hb_002639_030 |
0.0793781857 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 12 |
Hb_000418_020 |
0.0794728317 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_019654_020 |
0.0797341957 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 14 |
Hb_005854_040 |
0.0807845102 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002249_080 |
0.0814471298 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 16 |
Hb_116349_120 |
0.0841952719 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_000367_180 |
0.0884544056 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 18 |
Hb_000815_300 |
0.0885795725 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 19 |
Hb_002301_030 |
0.0891919787 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 20 |
Hb_001159_060 |
0.0895195915 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |