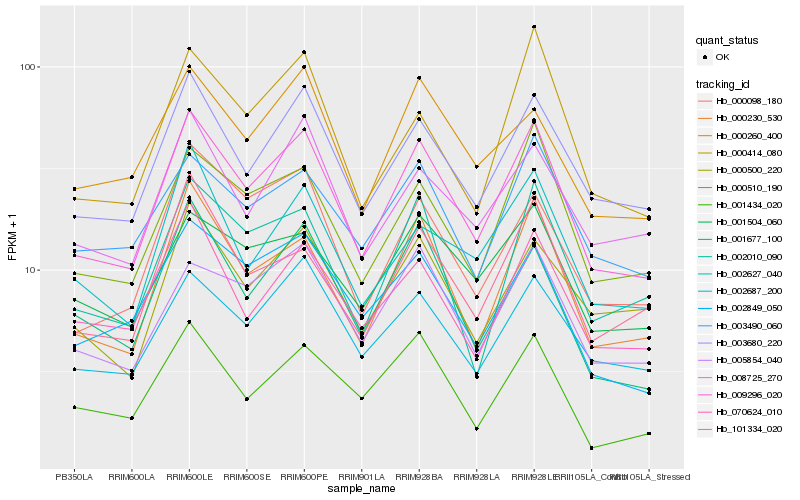
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009296_020 |
0.0 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_003680_220 |
0.0712942067 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 3 |
Hb_000230_530 |
0.0750016752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000510_190 |
0.0845119007 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 5 |
Hb_005854_040 |
0.0890901291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002687_200 |
0.0900646318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001677_100 |
0.0937687422 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 8 |
Hb_002849_050 |
0.0942038408 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 9 |
Hb_070624_010 |
0.0950395206 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 10 |
Hb_001434_020 |
0.0966376315 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 11 |
Hb_003490_060 |
0.0978316159 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 12 |
Hb_001504_060 |
0.0982084373 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 13 |
Hb_000098_180 |
0.0984518727 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 14 |
Hb_008725_270 |
0.0989479693 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 15 |
Hb_101334_020 |
0.0993401638 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 16 |
Hb_000500_220 |
0.0998075563 |
- |
- |
PREDICTED: intersectin-1 [Jatropha curcas] |
| 17 |
Hb_002010_090 |
0.1002282377 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000414_080 |
0.1010385934 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000260_400 |
0.1015252256 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 20 |
Hb_002627_040 |
0.1016435078 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |