| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
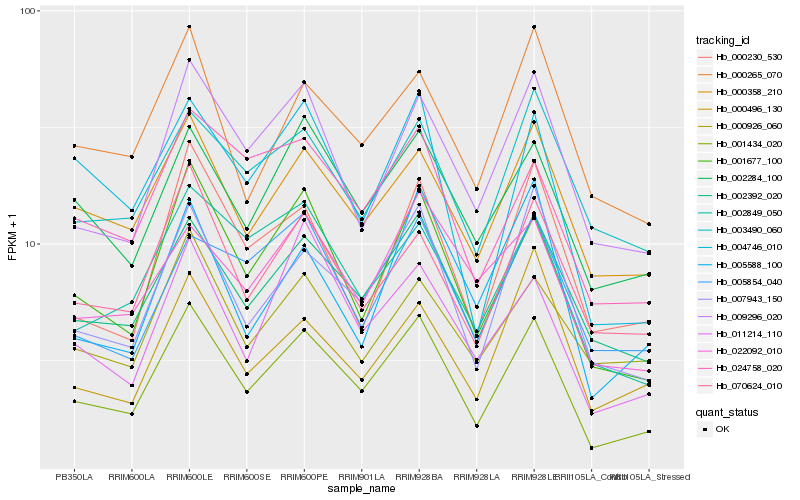
Hb_001434_020 |
0.0 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 2 |
Hb_001677_100 |
0.0808686949 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 3 |
Hb_002392_020 |
0.083942292 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 4 |
Hb_007943_150 |
0.0861588532 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_009296_020 |
0.0966376315 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_000358_210 |
0.0979156504 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 7 |
Hb_070624_010 |
0.1017366313 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 8 |
Hb_002284_100 |
0.1025523709 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 9 |
Hb_000230_530 |
0.1064719183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004746_010 |
0.1125941244 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 11 |
Hb_000265_070 |
0.1130505287 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 12 |
Hb_002849_050 |
0.113808054 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 13 |
Hb_024758_020 |
0.1152046696 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 14 |
Hb_003490_060 |
0.118417657 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 15 |
Hb_011214_110 |
0.11880677 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 16 |
Hb_000496_130 |
0.1215221482 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 17 |
Hb_000926_060 |
0.1216092942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_022092_010 |
0.1218271671 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 19 |
Hb_005854_040 |
0.1229428432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005588_100 |
0.1237252968 |
- |
- |
protein phosphatase, putative [Ricinus communis] |